Redoxins as gatekeepers of the transcriptional oxidative stress response
- PMID: 30690320
- PMCID: PMC6351230
- DOI: 10.1016/j.redox.2019.101104
Redoxins as gatekeepers of the transcriptional oxidative stress response
Abstract
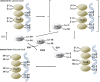
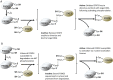
Transcription factors control the rate of transcription of genetic information from DNA to messenger RNA, by binding specific DNA sequences in promoter regions. Transcriptional gene control is a rate-limiting process that is tightly regulated and based on transient environmental signals which are translated into long-term changes in gene transcription. Post-translational modifications (PTMs) on transcription factors by phosphorylation or acetylation have profound effects not only on sub-cellular localization but also on substrate specificity through changes in DNA binding capacity. During times of cellular stress, specific transcription factors are in place to help protect the cell from damage by initiating the transcription of antioxidant response genes. Here we discuss PTMs caused by reactive oxygen species (ROS), such as H2O2, that can expeditiously regulate the activation of transcription factors involved in the oxidative stress response. Part of this rapid regulation are proteins involved in H2O2-related reduction and oxidation (redox) reactions such as redoxins, H2O2 scavengers described to interact with transcription factors. Redoxins have highly reactive cysteines of rate constants around 6-10-1 s-1 that engage in nucleophilic substitution of a thiol-disulfide with another thiol in inter-disulfide exchange reactions. We propose here that H2O2 signal transduction induced inter-disulfide exchange reactions between redoxin cysteines and cysteine thiols of transcription factors to allow for rapid and precise on and off switching of transcription factor activity. Thus, redoxins are essential modulators of stress response pathways beyond H2O2 scavenging capacity.
Copyright © 2019 The Authors. Published by Elsevier B.V. All rights reserved.
Figures



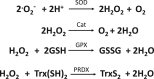
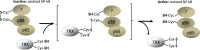
References
-
- Latchman D.S. Transcription factors: an overview. Int. J. Biochem. Cell Biol. 1997;29(12):1305–1312. - PubMed
-
- Gill G. Regulation of the initiation of eukaryotic transcription. Essays Biochem. 2001;37:33–43. - PubMed
-
- Xu L., Glass C.K., Rosenfeld M.G. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 1999;9(2):140–147. - PubMed
-
- Narlikar G.J., Fan H.Y., Kingston R.E. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108(4):475–487. - PubMed
-
- Vandromme M., Gauthier-Rouviere C., Lamb N., Fernandez A. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem. Sci. 1996;21(2):59–64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

