CRISPR-Cas9 mediated targeted disruption of FAD2-2 microsomal omega-6 desaturase in soybean (Glycine max.L)
- PMID: 30691438
- PMCID: PMC6350355
- DOI: 10.1186/s12896-019-0501-2
CRISPR-Cas9 mediated targeted disruption of FAD2-2 microsomal omega-6 desaturase in soybean (Glycine max.L)
Erratum in
-
Correction to: CRISPR-Cas9 mediated targeted disruption of FAD2-2 microsomal omega-6 desaturase in soybean (Glycine max.L).BMC Biotechnol. 2020 Aug 20;20(1):42. doi: 10.1186/s12896-020-00634-x. BMC Biotechnol. 2020. PMID: 32819342 Free PMC article.
Abstract
Background: Recent innovation in the field of genome engineering encompasses numerous levels of plant genome engineering which attract the substantial excitement of plant biologist worldwide. RNA-guided CRISPR Cas9 system has appeared a promising tool in site-directed mutagenesis due to its innovative utilization in different branches of biology. CRISPR-Cas9 nuclease system have supersedes all previously existed strategies and their associated pitfalls encountered with site-specific mutagenesis.
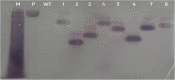
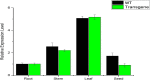
Results: Here we demonstrated an efficient sequence specific integration/mutation of FAD2-2 gene in soybean using CRISPR-Cas9 nuclease system. A single guided RNA sequence was designed with the help of a number of bioinformatics tools aimed to target distinct sites of FAD2-2 loci in soybean. The binary vector (pCas9-AtU6-sgRNA) has been successfully transformed into soybean cotyledon using Agrobacterium tumafacien. Taken together our findings complies soybean transgenic mutants subjected to targeted mutation were surprisingly detected in our target gene. Furthermore, the detection of Cas9 gene, BAR gene, and NOS terminator were carried out respectively. Southern blot analysis confirmed the stable transformation of Cas9 gene into soybean. Real time expression with qRT-PCR and Sanger sequencing analysis confirmed the efficient CRISPR-Cas9/sgRNA induced mutation within the target sequence of FAD2-2 loci. The integration of FAD2-2 target region in the form of substitution, deletions and insertions were achieved with notably high frequency and rare off-target mutagenesis.
Conclusion: High frequent mutation efficiency was recorded as 21% out of all transgenic soybean plants subjected to targeted mutagenesis. Furthermore, Near-infrared spectroscopy (NIR) indicates the entire fatty acid profiling obtained from the mutants seeds of soybean. A considerable modulation in oleic acid content up to (65.58%) whereas the least level of linoleic acid is (16.08%) were recorded. Based on these finding CRISPR-Cas9 system can possibly sum up recent development and future challenges in producing agronomically important crops.
Keywords: CRISPR-Cas9; FAD2–2; Oleic acid; Soybean; Targeted mutagenesis.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
-
- Girija M, Dhanavel D, Gnanamurthy S. Gamma Rays and EMS Induced Flower Color and Seed Mutants in Cowpea (Vignaunguiculata L. Walp) Advances in Applied Science Research. 2013;4:134–139.
-
- Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2014;111:4632–4637. doi: 10.1073/pnas.1400822111. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

