Kinetic characterization of Cas9 enzymes
- PMID: 30691648
- PMCID: PMC8364395
- DOI: 10.1016/bs.mie.2018.10.022
Kinetic characterization of Cas9 enzymes
Abstract
Bacterial adaptive immune systems employ clustered regularly interspaced short palindromic repeats (CRISPR) along with their CRISPR-associated genes (Cas) to form CRISPR RNA (crRNA)-guided surveillance complexes, which target foreign nucleic acids for destruction. Cas9 is unique in that it is composed of a single polypeptide that utilizes both a crRNA and a trans-activating crRNA (tracrRNA) or a single guide RNA to create double-stranded breaks in sequences complementary to the RNA via the HNH and RuvC nuclease domains. Cas9 has become a revolutionary tool for gene-editing applications. Here, we describe methods for studying the cleavage activities of Cas9. We describe protocols for rapid quench-flow and stopped-flow kinetics and interpretation of the results. The protocols detailed here will be paramount for understanding the mechanistic basis for specificity of this enzyme, especially in efforts to improve accuracy for clinical use.
Keywords: CRISPR; Cas9; Enzyme kinetics; Genome editing.
© 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Financial conflict of interest statement: KAJ is President of KinTek Corporation, which provided the stopped-flow and quench-flow instruments and KinTek Explorer software used in this study.
Figures

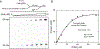

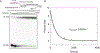



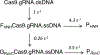
References
-
- Doudna JA and Charpentier E (2014) ‘Genome editing. The new frontier of genome engineering with CRISPR-Cas9’, Science, 346(6213), pp. 1258096. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

