Zfp423 Regulates Skeletal Muscle Regeneration and Proliferation
- PMID: 30692273
- PMCID: PMC6447414
- DOI: 10.1128/MCB.00447-18
Zfp423 Regulates Skeletal Muscle Regeneration and Proliferation
Abstract
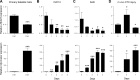
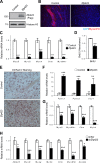
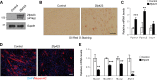
Satellite cells (SCs) are skeletal muscle stem cells that proliferate in response to injury and provide myogenic precursors for growth and repair. Zfp423 is a transcriptional cofactor expressed in multiple immature cell populations, such as neuronal precursors, mesenchymal stem cells, and preadipocytes, where it regulates lineage allocation, proliferation, and differentiation. Here, we show that Zfp423 regulates myogenic progression during muscle regeneration. Zfp423 is undetectable in quiescent SCs but becomes expressed during SC activation. After expansion, Zfp423 is gradually downregulated as committed SCs terminally differentiate. Mice with satellite-cell-specific Zfp423 deletion exhibit severely impaired muscle regeneration following injury, with aberrant SC expansion, defective cell cycle exit, and failure to transition efficiently from the proliferative stage toward commitment. Consistent with a cell-autonomous role of Zfp423, shRNA-mediated knockdown of Zfp423 in myoblasts inhibits differentiation. Surprisingly, forced expression of Zfp423 in myoblasts induces differentiation into adipocytes and arrests myogenesis. Affinity purification of Zfp423 in myoblasts identified Satb2 as a nuclear partner of Zfp423 that cooperatively enhances Zfp423 transcriptional activity, which in turn affects myoblast differentiation. In conclusion, by controlling SC expansion and proliferation, Zfp423 is essential for muscle regeneration. Tight regulation of Zfp423 expression is essential for normal progression of muscle progenitors from proliferation to differentiation.
Keywords: cell fate; myoblast; myogenesis; regeneration; satellite cells; skeletal muscle.
Copyright © 2019 Addison et al.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
