Both anti-inflammatory and antiviral properties of novel drug candidate ABX464 are mediated by modulation of RNA splicing
- PMID: 30692590
- PMCID: PMC6349857
- DOI: 10.1038/s41598-018-37813-y
Both anti-inflammatory and antiviral properties of novel drug candidate ABX464 are mediated by modulation of RNA splicing
Abstract
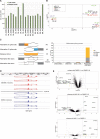
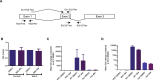
ABX464 is a first-in-class, clinical-stage, small molecule for oral administration that has shown strong anti-inflammatory effects in the DSS-model for inflammatory bowel disease (IBD) and also prevents replication of the HIV virus. ABX464 which binds to cap binding complex (CBC) has demonstrated safety and efficacy in a phase 2a proof-of-concept clinical trial in patients with Ulcerative colitis. Previously, with limited technologies, it was not possible to quantify the effect of ABX464 on viral and cellular RNA biogenesis. Here, using RNA CaptureSeq and deep sequencing, we report that ABX464 enhances the splicing of HIV RNA in infected PBMCs from six healthy individuals and also the expression and splicing of a single long noncoding RNA to generate the anti-inflammatory miR-124 both ex vivo and in HIV patients. While ABX464 has no effect on pre-mRNA splicing of cellular genes, depletion of CBC complex by RNAi leads to accumulation of intron retention transcripts. These results imply that ABX464 did not inhibit the function of CBC in splicing but rather strengthens it under pathological condition like inflammation and HIV infection. The specific dual ability of ABX464 to generate both anti-inflammatory miR-124 and spliced viral RNA may have applicability for the treatment of both inflammatory diseases and HIV infection.
Conflict of interest statement
A.V., D.S., A.G., N.C. H.E. and J.T. are Abivax employees. D.S., H.E. and J.T. hold Abivax stock options. J.T. is member of ABIVAX SAB and the collaborative laboratory that has received financial support from Abivax.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

