Sequentially inducible mouse models reveal that Npm1 mutation causes malignant transformation of Dnmt3a-mutant clonal hematopoiesis
- PMID: 30692594
- PMCID: PMC6609470
- DOI: 10.1038/s41375-018-0368-6
Sequentially inducible mouse models reveal that Npm1 mutation causes malignant transformation of Dnmt3a-mutant clonal hematopoiesis
Abstract
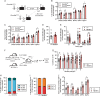
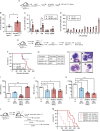
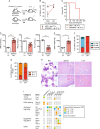
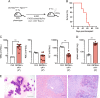
Clonal hematopoiesis (CH) is a common aging-associated condition with increased risk of hematologic malignancy. Knowledge of the mechanisms driving evolution from CH to overt malignancy has been hampered by a lack of in vivo models that orthogonally activate mutant alleles. Here, we develop independently regulatable mutations in DNA methyltransferase 3A (Dnmt3a) and nucleophosmin 1 (Npm1), observed in human CH and AML, respectively. We find Dnmt3a mutation expands hematopoietic stem and multipotent progenitor cells (HSC/MPPs), modeling CH. Induction of mutant Npm1 after development of Dnmt3a-mutant CH causes progression to myeloproliferative disorder (MPD), and more aggressive MPD is observed with longer latency between mutations. MPDs uniformly progress to acute myeloid leukemia (AML) following transplant, accompanied by a decrease in HSC/MPPs and an increase in myeloid-restricted progenitors, the latter of which propagate AML in tertiary recipient mice. At a molecular level, progression of CH to MPD is accompanied by selection for mutations activating Ras/Raf/MAPK signaling. Progression to AML is characterized by additional oncogenic signaling mutations (Ptpn11, Pik3r1, Flt3) and/or mutations in epigenetic regulators (Hdac1, Idh1, Arid1a). Together, our study demonstrates that Npm1 mutation drives evolution of Dnmt3a-mutant CH to AML and rate of disease progression is accelerated with longer latency of CH.
Conflict of interest statement
R.L. is on the supervisory board of Qiagen and is a scientific advisor to Loxo, Imago, C4 Therapeutics, and Isoplexis. He receives research support from and consulted for Celgene and Roche, research support from Prelude Therapeutics, and has consulted for Novartis and Gilead. He has received honoraria from Lilly and Amgen for invited lectures. The remaining authors declare that they have no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

