Bacterial sensors define intracellular free energies for correct enzyme metalation
- PMID: 30692683
- PMCID: PMC6420079
- DOI: 10.1038/s41589-018-0211-4
Bacterial sensors define intracellular free energies for correct enzyme metalation
Abstract
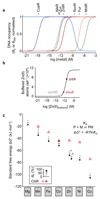
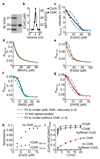
There is a challenge for metalloenzymes to acquire their correct metals because some inorganic elements form more stable complexes with proteins than do others. These preferences can be overcome provided some metals are more available than others. However, while the total amount of cellular metal can be readily measured, the available levels of each metal have been more difficult to define. Metal-sensing transcriptional regulators are tuned to the intracellular availabilities of their cognate ions. Here we have determined the standard free energy for metal complex formation to which each sensor, in a set of bacterial metal sensors, is attuned: the less competitive the metal, the less favorable the free energy and hence the greater availability to which the cognate allosteric mechanism is tuned. Comparing these free energies with values derived from the metal affinities of a metalloprotein reveals the mechanism of correct metalation exemplified here by a cobalt chelatase for vitamin B12.
Conflict of interest statement
Figures



References
-
- Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. - PubMed
-
- Tottey S, et al. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature. 2008;455:1138–1142. - PubMed
-
- Fraústo da Silva JJR, Williams RJP. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. Oxford University Press; Oxford: 1991.
-
- Irving H, Williams RJP. Order of stability of metal complexes. Nature. 1948;162:746–747.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

