Identification of preexisting adaptive immunity to Cas9 proteins in humans
- PMID: 30692695
- PMCID: PMC7199589
- DOI: 10.1038/s41591-018-0326-x
Identification of preexisting adaptive immunity to Cas9 proteins in humans
Abstract
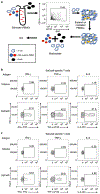
The CRISPR-Cas9 system is a powerful tool for genome editing, which allows the precise modification of specific DNA sequences. Many efforts are underway to use the CRISPR-Cas9 system to therapeutically correct human genetic diseases1-6. The most widely used orthologs of Cas9 are derived from Staphylococcus aureus and Streptococcus pyogenes5,7. Given that these two bacterial species infect the human population at high frequencies8,9, we hypothesized that humans may harbor preexisting adaptive immune responses to the Cas9 orthologs derived from these bacterial species, SaCas9 (S. aureus) and SpCas9 (S. pyogenes). By probing human serum for the presence of anti-Cas9 antibodies using an enzyme-linked immunosorbent assay, we detected antibodies against both SaCas9 and SpCas9 in 78% and 58% of donors, respectively. We also found anti-SaCas9 T cells in 78% and anti-SpCas9 T cells in 67% of donors, which demonstrates a high prevalence of antigen-specific T cells against both orthologs. We confirmed that these T cells were Cas9-specific by demonstrating a Cas9-specific cytokine response following isolation, expansion, and antigen restimulation. Together, these data demonstrate that there are preexisting humoral and cell-mediated adaptive immune responses to Cas9 in humans, a finding that should be taken into account as the CRISPR-Cas9 system moves toward clinical trials.
Figures



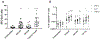






Comment in
-
Human immunity to Cas9.Nat Methods. 2019 Apr;16(4):286. doi: 10.1038/s41592-019-0385-z. Nat Methods. 2019. PMID: 30923392 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

