Enzyme-Enzyme Interactions in Monolignol Biosynthesis
- PMID: 30693007
- PMCID: PMC6340093
- DOI: 10.3389/fpls.2018.01942
Enzyme-Enzyme Interactions in Monolignol Biosynthesis
Abstract
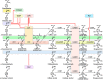
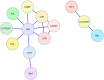
The enzymes that comprise the monolignol biosynthetic pathway have been studied intensively for more than half a century. A major interest has been the role of pathway in the biosynthesis of lignin and the role of lignin in the formation of wood. The pathway has been typically conceived as linear steps that convert phenylalanine into three major monolignols or as a network of enzymes in a metabolic grid. Potential interactions of enzymes have been investigated to test models of metabolic channeling or for higher order interactions. Evidence for enzymatic or physical interactions has been fragmentary and limited to a few enzymes studied in different species. Only recently the entire pathway has been studied comprehensively in any single plant species. Support for interactions comes from new studies of enzyme activity, co-immunoprecipitation, chemical crosslinking, bimolecular fluorescence complementation, yeast 2-hybrid functional screening, and cell type-specific gene expression based on light amplification by stimulated emission of radiation capture microdissection. The most extensive experiments have been done on differentiating xylem of Populus trichocarpa, where genomic, biochemical, chemical, and cellular experiments have been carried out. Interactions affect the rate, direction, and specificity of both 3 and 4-hydroxylation in the monolignol biosynthetic pathway. Three monolignol P450 mono-oxygenases form heterodimeric and heterotetrameric protein complexes that activate specific hydroxylation of cinnamic acid derivatives. Other interactions include regulatory kinetic control of 4-coumarate CoA ligases through subunit specificity and interactions between a cinnamyl alcohol dehydrogenase and a cinnamoyl-CoA reductase. Monolignol enzyme interactions with other pathway proteins have been associated with biotic and abiotic stress response. Evidence challenging or supporting metabolic channeling in this pathway will be discussed.
Keywords: BiFC; co-immunoprecipitation; enzyme kinetics; lignin; monolignol biosynthesis; protein-protein interaction.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources

