Deducting MicroRNA-Mediated Changes Common in Bronchial Epithelial Cells of Asthma and Chronic Obstructive Pulmonary Disease-A Next-Generation Sequencing-Guided Bioinformatic Approach
- PMID: 30696075
- PMCID: PMC6386886
- DOI: 10.3390/ijms20030553
Deducting MicroRNA-Mediated Changes Common in Bronchial Epithelial Cells of Asthma and Chronic Obstructive Pulmonary Disease-A Next-Generation Sequencing-Guided Bioinformatic Approach
Abstract
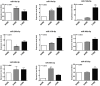
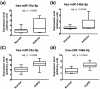
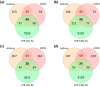
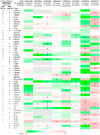
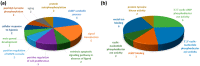
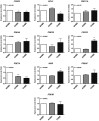
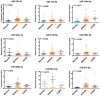
Asthma and chronic obstructive pulmonary disease (COPD) are chronic airway inflammatory diseases that share some common features, although these diseases are somewhat different in etiologies, clinical features, and treatment policies. The aim of this study is to investigate the common microRNA-mediated changes in bronchial epithelial cells of asthma and COPD. The microRNA profiles in primary bronchial epithelial cells from asthma (AHBE) and COPD (CHBE) patients and healthy subjects (NHBE) were analyzed with next-generation sequencing (NGS) and the significant microRNA changes common in AHBE and CHBE were extracted. The upregulation of hsa-miR-10a-5p and hsa-miR-146a-5p in both AHBE and CHBE was confirmed with quantitative polymerase chain reaction (qPCR). Using bioinformatic methods, we further identified putative targets of these microRNAs, which were downregulated in both AHBE and CHBE: miR-10a-5p might suppress BCL2, FGFR3, FOXO3, PDE4A, PDE4C, and PDE7A; miR-146a-5p might suppress BCL2, INSR, PDE4D, PDE7A, PDE7B, and PDE11A. We further validated significantly decreased expression levels of FOXO3 and PDE7A in AHBE and CHBE than in NHBE with qPCR. Increased serum miR-146a-5p level was also noted in patients with asthma and COPD as compared with normal control subjects. In summary, our study revealed possible mechanisms mediated by miR-10a-5p and miR-146a-5p in the pathogenesis of both asthma and COPD. The findings might provide a scientific basis for developing novel diagnostic and therapeutic strategies.
Keywords: COPD; asthma; bioinformatics; bronchial epithelial cells; epithelium; next-generation sequencing.
Conflict of interest statement
The authors declare no related conflict of interest.
Figures
Similar articles
-
Possible mechanisms mediating apoptosis of bronchial epithelial cells in chronic obstructive pulmonary disease - A next-generation sequencing approach.Pathol Res Pract. 2018 Sep;214(9):1489-1496. doi: 10.1016/j.prp.2018.08.002. Epub 2018 Aug 7. Pathol Res Pract. 2018. PMID: 30115538
-
MicroRNA Profiling Reveals a Role for MicroRNA-218-5p in the Pathogenesis of Chronic Obstructive Pulmonary Disease.Am J Respir Crit Care Med. 2017 Jan 1;195(1):43-56. doi: 10.1164/rccm.201506-1182OC. Am J Respir Crit Care Med. 2017. PMID: 27409149
-
The Plasma Levels of hsa-miR-19b-3p, hsa-miR-125b-5p, and hsamiR- 320c in Patients with Asthma, COPD and Asthma-COPD Overlap Syndrome (ACOS).Microrna. 2021;10(2):130-138. doi: 10.2174/2211536610666210609142859. Microrna. 2021. PMID: 34151771
-
MicroRNAs: future biomarkers and targets of therapy in asthma?Curr Opin Pulm Med. 2020 May;26(3):285-292. doi: 10.1097/MCP.0000000000000673. Curr Opin Pulm Med. 2020. PMID: 32101904 Review.
-
miR-223: A Key Regulator in the Innate Immune Response in Asthma and COPD.Front Med (Lausanne). 2020 May 19;7:196. doi: 10.3389/fmed.2020.00196. eCollection 2020. Front Med (Lausanne). 2020. PMID: 32509795 Free PMC article. Review.
Cited by
-
Emerging Advances of Non-coding RNAs and Competitive Endogenous RNA Regulatory Networks in Asthma.Bioengineered. 2021 Dec;12(1):7820-7836. doi: 10.1080/21655979.2021.1981796. Bioengineered. 2021. PMID: 34635022 Free PMC article. Review.
-
miR-150-5p-Containing Extracellular Vesicles Are a New Immunoregulator That Favor the Progression of Lung Cancer in Hypoxic Microenvironments by Altering the Phenotype of NK Cells.Cancers (Basel). 2021 Dec 13;13(24):6252. doi: 10.3390/cancers13246252. Cancers (Basel). 2021. PMID: 34944871 Free PMC article.
-
Transcriptomic analysis reveals pathophysiological relationship between chronic obstructive pulmonary disease (COPD) and periodontitis.BMC Med Genomics. 2022 Jun 8;15(1):130. doi: 10.1186/s12920-022-01278-w. BMC Med Genomics. 2022. PMID: 35676670 Free PMC article.
-
Dual role of the miR-146 family in rhinovirus-induced airway inflammation and allergic asthma exacerbation.Clin Transl Med. 2021 Jun;11(6):e427. doi: 10.1002/ctm2.427. Clin Transl Med. 2021. PMID: 34185416 Free PMC article.
-
Reduced expression of miR-146a in human bronchial epithelial cells alters neutrophil migration.Clin Transl Allergy. 2019 Nov 27;9:62. doi: 10.1186/s13601-019-0301-8. eCollection 2019. Clin Transl Allergy. 2019. PMID: 31798831 Free PMC article.
References
-
- Sheu C.C., Tsai M.J., Chen F.W., Chang K.F., Chang W.A., Chong I.W., Kuo P.L., Hsu Y.L. Identification of novel genetic regulations associated with airway epithelial homeostasis using next-generation sequencing data and bioinformatics approaches. Oncotarget. 2017;8:82674–82688. doi: 10.18632/oncotarget.19752. - DOI - PMC - PubMed
-
- Tsai M.J., Chang W.A., Jian S.F., Chang K.F., Sheu C.C., Kuo P.L. Possible mechanisms mediating apoptosis of bronchial epithelial cells in chronic obstructive pulmonary disease—A next-generation sequencing approach. Pathol. Res. Pract. 2018;214:1489–1496. doi: 10.1016/j.prp.2018.08.002. - DOI - PubMed
-
- GINA . Global Strategy for Asthma Management and Prevention. Global Initiative for Asthma (GINA); Fontana, WI, USA: 2018.
-
- GOLD . Global Strategy for Diagnosis, Management, and Prevention of COPD. Global Initiative for Obstructive Lung Disease (GOLD); 2018. [(accessed on 28 January 2019)]. Available online: https://goldcopd.org/
MeSH terms
Substances
Grants and funding
- NSC 102-2220-E-037-001/National Science Council
- MOST 104-2314-B-037-034-MY3/Ministry of Science and Technology, Taiwan
- MOST 107-2320-B-037-011-MY3/Ministry of Science and Technology, Taiwan
- KMUHS10601/Kaohsiung Medical University Chung-Ho Memorial Hospital
- KMUH106-6R12/Kaohsiung Medical University Chung-Ho Memorial Hospital
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

