GPR68: An Emerging Drug Target in Cancer
- PMID: 30696114
- PMCID: PMC6386835
- DOI: 10.3390/ijms20030559
GPR68: An Emerging Drug Target in Cancer
Abstract
GPR68 (or ovarian cancer G protein-coupled receptor 1, OGR1) is a proton-sensing G-protein-coupled receptor (GPCR) that responds to extracellular acidity and regulates a variety of cellular functions. Acidosis is considered a defining hallmark of the tumor microenvironment (TME). GPR68 expression is highly upregulated in numerous types of cancer. Emerging evidence has revealed that GPR68 may play crucial roles in tumor biology, including tumorigenesis, tumor growth, and metastasis. This review summarizes current knowledge regarding GPR68-its expression, regulation, signaling pathways, physiological roles, and functions it regulates in human cancers (including prostate, colon and pancreatic cancer, melanoma, medulloblastoma, and myelodysplastic syndrome). The findings provide evidence for GPR68 as a potentially novel therapeutic target but in addition, we note challenges in developing drugs that target GPR68.
Keywords: GPR68; RNA-seq; acidosis; proton-sensing GPCRs; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

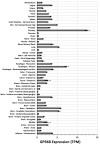
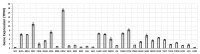
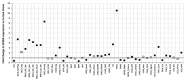
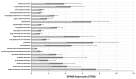

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

