IDH mutation-specific radiomic signature in lower-grade gliomas
- PMID: 30696801
- PMCID: PMC6366985
- DOI: 10.18632/aging.101769
IDH mutation-specific radiomic signature in lower-grade gliomas
Abstract
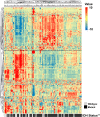
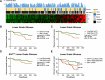
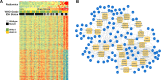
Unravelling the heterogeneity is the central challenge for glioma precession oncology. In this study, we extracted quantitative image features from T2-weighted MR images and revealed that the isocitrate dehydrogenase (IDH) wild type and mutant lower grade gliomas (LGGs) differed in their expression of 146 radiomic descriptors. The logistic regression model algorithm further reduced these to 86 features. The classification model could discriminate the two types in both the training and validation sets with area under the curve values of 1.0000 and 0.9932, respectively. The transcriptome-radiomic analysis revealed that these features were associated with the immune response, biological adhesion, and several malignant behaviors, all of which are consistent with biological processes that are differentially expressed in IDH wild type and IDH mutant LGGs. Finally, a prognostic signature showed an ability to stratify IDH mutant LGGs into high and low risk groups with distinctive outcomes. By extracting a large number of radiomic features, we identified an IDH mutation-specific radiomic signature with prognostic implications. This radiomic signature may provide a way to non-invasively discriminate lower-grade gliomas as with or without the IDH mutation.
Keywords: isocitrate dehydrogenase; lower grade gliomas; prognostic signature; radiomic signature; transcriptome-radiomic analysis.
Conflict of interest statement
Figures



References
-
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta neuropathologica. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

