Disentangling the complexity of low complexity proteins
- PMID: 30698641
- PMCID: PMC7299295
- DOI: 10.1093/bib/bbz007
Disentangling the complexity of low complexity proteins
Abstract
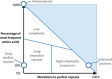
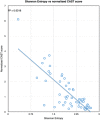
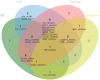
There are multiple definitions for low complexity regions (LCRs) in protein sequences, with all of them broadly considering LCRs as regions with fewer amino acid types compared to an average composition. Following this view, LCRs can also be defined as regions showing composition bias. In this critical review, we focus on the definition of sequence complexity of LCRs and their connection with structure. We present statistics and methodological approaches that measure low complexity (LC) and related sequence properties. Composition bias is often associated with LC and disorder, but repeats, while compositionally biased, might also induce ordered structures. We illustrate this dichotomy, and more generally the overlaps between different properties related to LCRs, using examples. We argue that statistical measures alone cannot capture all structural aspects of LCRs and recommend the combined usage of a variety of predictive tools and measurements. While the methodologies available to study LCRs are already very advanced, we foresee that a more comprehensive annotation of sequences in the databases will enable the improvement of predictions and a better understanding of the evolution and the connection between structure and function of LCRs. This will require the use of standards for the generation and exchange of data describing all aspects of LCRs.
Short abstract: There are multiple definitions for low complexity regions (LCRs) in protein sequences. In this critical review, we focus on the definition of sequence complexity of LCRs and their connection with structure. We present statistics and methodological approaches that measure low complexity (LC) and related sequence properties. Composition bias is often associated with LC and disorder, but repeats, while compositionally biased, might also induce ordered structures. We illustrate this dichotomy, plus overlaps between different properties related to LCRs, using examples.
Keywords: composition bias; disorder; low complexity regions; structure.
© The Author(s) 2019. Published by Oxford University Press.
Figures



References
-
- Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys 2008;37:215–246. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

