C/EBPβ suppresses keratinocyte autonomous type 1 IFN response and p53 to increase cell survival and susceptibility to UVB-induced skin cancer
- PMID: 30698678
- PMCID: PMC10893916
- DOI: 10.1093/carcin/bgz012
C/EBPβ suppresses keratinocyte autonomous type 1 IFN response and p53 to increase cell survival and susceptibility to UVB-induced skin cancer
Abstract
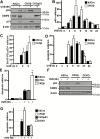
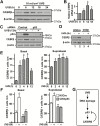
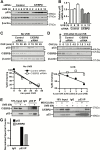
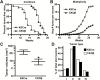
p53 is activated by DNA damage and oncogenic stimuli to regulate senescence, apoptosis and cell-cycle arrest, which are essential to prevent cancer. Here, we utilized UVB radiation, a potent inducer of DNA damage, p53, apoptosis and skin cancer to investigate the mechanism of CCAAT/enhancer binding protein-β (C/EBPβ) in regulating p53-mediated apoptosis in keratinocytes and to test whether the deletion of C/EBPβ in epidermis can protect mice from UVB-induced skin cancer. UVB-treatment of C/EBPβ skin conditional knockout (CKOβ) mice increased p53 protein levels in epidermis and enhanced p53-dependent apoptotic activity 3-fold compared with UVB-treated control mice. UVB increased C/EBPβ levels through a p53-dependent pathway and stimulated the formation of a C/EBPβ-p53 protein complex; knockdown of C/EBPβ increased p53 protein stability in keratinocytes. These results suggest a p53-C/EBPβ feedback loop, whereby C/EBPβ, a transcriptional target of a p53 pathway, functions as a survival factor by negatively regulating p53 apoptotic activity in response to DNA damage. RNAseq analysis of UVB-treated CKOβ epidermis unexpectedly revealed that type 1 interferon (IFN) pathway was the most highly enriched pathway. Numerous pro-apoptotic interferon stimulated genes were upregulated including some known to enhance p53 apoptosis. Our results indicate that p53 and IFN pathways function together in response to DNA damage to result in the activation of extrinsic apoptosis pathways and caspase 8 cleavage. Last, we observed CKOβ mice were resistant to UVB-induced skin cancer. Our results suggest that C/EBPβ represses apoptosis through keratinocyte autonomous suppression of the type 1 IFN response and p53 to increase cell survival and susceptibility to UVB-induced skin cancer.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

