The y-ome defines the 35% of Escherichia coli genes that lack experimental evidence of function
- PMID: 30698741
- PMCID: PMC6412132
- DOI: 10.1093/nar/gkz030
The y-ome defines the 35% of Escherichia coli genes that lack experimental evidence of function
Abstract
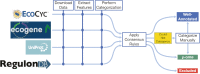
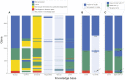
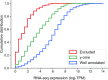
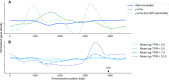
Experimental studies of Escherichia coli K-12 MG1655 often implicate poorly annotated genes in cellular phenotypes. However, we lack a systematic understanding of these genes. How many are there? What information is available for them? And what features do they share that could explain the gap in our understanding? Efforts to build predictive, whole-cell models of E. coli inevitably face this knowledge gap. We approached these questions systematically by assembling annotations from the knowledge bases EcoCyc, EcoGene, UniProt and RegulonDB. We identified the genes that lack experimental evidence of function (the 'y-ome') which include 1600 of 4623 unique genes (34.6%), of which 111 have absolutely no evidence of function. An additional 220 genes (4.7%) are pseudogenes or phantom genes. y-ome genes tend to have lower expression levels and are enriched in the termination region of the E. coli chromosome. Where evidence is available for y-ome genes, it most often points to them being membrane proteins and transporters. We resolve the misconception that a gene in E. coli whose primary name starts with 'y' is unannotated, and we discuss the value of the y-ome for systematic improvement of E. coli knowledge bases and its extension to other organisms.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Hutchison C.A. 3rd, Chuang R.-Y., Noskov V.N., Assad-Garcia N., Deerinck T.J., Ellisman M.H., Gill J., Kannan K., Karas B.J., Ma L. et al.. Design and synthesis of a minimal bacterial genome. Science. 2016; 351:aad6253. - PubMed
-
- Dellomonaco C., Clomburg J.M., Miller E.N., Gonzalez R.. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature. 2011; 476:355–359. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

