Kinetic analysis of N-alkylaryl carboxamide hexitol nucleotides as substrates for evolved polymerases
- PMID: 30698800
- PMCID: PMC6412122
- DOI: 10.1093/nar/gkz008
Kinetic analysis of N-alkylaryl carboxamide hexitol nucleotides as substrates for evolved polymerases
Abstract
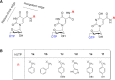
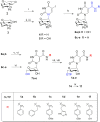
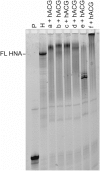
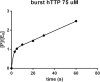
Six 1',5'-anhydrohexitol uridine triphosphates were synthesized with aromatic substitutions appended via a carboxamide linker to the 5-position of their bases. An improved method for obtaining such 5-substituted hexitol nucleosides and nucleotides is described. The incorporation profile of the nucleotide analogues into a DNA duplex overhang using recently evolved XNA polymerases is compared. Long, mixed HNA sequences featuring the base modifications are generated. The apparent binding affinity of four of the nucleotides to the enzyme, the rate of the chemical step and of product release, plus the specificity constant for the incorporation of these modified nucleotides into a DNA duplex overhang using the HNA polymerase T6G12_I521L are determined via pre-steady-state kinetics. HNA polymers displaying aromatic functional groups could have significant impact on the isolation of stable and high-affinity binders and catalysts, or on the design of nanomaterials.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Enzymatic incorporation in DNA of 1,5-anhydrohexitol nucleotides.Biochemistry. 2000 Oct 24;39(42):12757-65. doi: 10.1021/bi001297g. Biochemistry. 2000. PMID: 11041840
-
Recognition of HNA and 1,5-anhydrohexitol nucleotides by DNA metabolizing enzymes.Biochim Biophys Acta. 2002 May 20;1597(1):115-22. doi: 10.1016/s0167-4838(02)00267-4. Biochim Biophys Acta. 2002. PMID: 12009410
-
Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.Biomolecules. 2020 Dec 8;10(12):1647. doi: 10.3390/biom10121647. Biomolecules. 2020. PMID: 33302546 Free PMC article.
-
Cross-coupling reactions of nucleoside triphosphates followed by polymerase incorporation. Construction and applications of base-functionalized nucleic acids.Org Biomol Chem. 2008 Jul 7;6(13):2233-41. doi: 10.1039/b803664k. Epub 2008 Apr 2. Org Biomol Chem. 2008. PMID: 18563253 Review.
-
A reexamination of the nucleotide incorporation fidelity of DNA polymerases.Biochemistry. 2002 Aug 27;41(34):10571-6. doi: 10.1021/bi026021i. Biochemistry. 2002. PMID: 12186540 Review.
Cited by
-
Modified nucleic acids: replication, evolution, and next-generation therapeutics.BMC Biol. 2020 Sep 2;18(1):112. doi: 10.1186/s12915-020-00803-6. BMC Biol. 2020. PMID: 32878624 Free PMC article. Review.
-
Efficient synthesis and replication of diverse sequence libraries composed of biostable nucleic acid analogues.RSC Chem Biol. 2022 Aug 30;3(10):1209-1215. doi: 10.1039/d2cb00035k. eCollection 2022 Oct 5. RSC Chem Biol. 2022. PMID: 36320888 Free PMC article.
-
Mononucleotide repeat expansions with non-natural polymerase substrates.Sci Rep. 2021 Jan 28;11(1):2423. doi: 10.1038/s41598-021-82150-2. Sci Rep. 2021. PMID: 33510377 Free PMC article.
-
Modified Nucleotides for Chemical and Enzymatic Synthesis of Therapeutic RNA.Curr Med Chem. 2023;30(11):1320-1347. doi: 10.2174/0929867330666221014111403. Curr Med Chem. 2023. PMID: 36239720
References
-
- Le B.T., Chen S., Abramov M., Herdewijn P., Veedu R.N.. Evaluation of anhydrohexitol nucleic acid, cyclohexenyl nucleic acid and d-altritol nucleic acid-modified 2′-O-methyl RNA mixmer antisense oligonucleotides for exon skipping in vitro. Chem. Commun. (Camb.). 2016; 52:13467–13470. - PubMed
-
- Pezo V., Liu F.W., Abramov M., Froeyen M., Herdewijn P., Marliere P.. Binary genetic cassettes for selecting XNA-templated DNA synthesis in vivo. Angew. Chem. Int. Ed. Engl. 2013; 52:8139–8143. - PubMed
-
- Pezo V., Schepers G., Lambertucci C., Marliere P., Herdewijn P.. Probing ambiguous base-pairs by genetic transformation with XNA templates. ChemBioChem. 2014; 15:2255–2258. - PubMed
-
- Pochet S., Kaminski P.A., Van Aerschot A., Herdewijn P., Marlière P.. Replication of hexitol oligonucleotides as a prelude to the propagation of a third type of nucleic acid in vivo. C. R. Biol. 2003; 326:1175–1184. - PubMed

