Antimicrobial Microwebs of DNA-Histone Inspired from Neutrophil Extracellular Traps
- PMID: 30698844
- PMCID: PMC6467213
- DOI: 10.1002/adma.201807436
Antimicrobial Microwebs of DNA-Histone Inspired from Neutrophil Extracellular Traps
Abstract
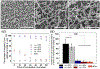
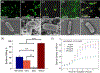
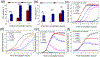
Neutrophil extracellular traps (NETs) are decondensed chromatin networks released by neutrophils that can trap and kill pathogens but can also paradoxically promote biofilms. The mechanism of NET functions remains ambiguous, at least in part, due to their complex and variable compositions. To unravel the antimicrobial performance of NETs, a minimalistic NET-like synthetic structure, termed "microwebs," is produced by the sonochemical complexation of DNA and histone. The prepared microwebs have structural similarity to NETs at the nanometer to micrometer dimensions but with well-defined molecular compositions. Microwebs prepared with different DNA to histone ratios show that microwebs trap pathogenic Escherichia coli in a manner similar to NETs when the zeta potential of the microwebs is positive. The DNA nanofiber networks and the bactericidal histone constituting the microwebs inhibit the growth of E. coli. Moreover, microwebs work synergistically with colistin sulfate, a common and a last-resort antibiotic, by targeting the cell envelope of pathogenic bacteria. The synthesis of microwebs enables mechanistic studies not possible with NETs, and it opens new possibilities for constructing biomimetic bacterial microenvironments to better understand and predict physiological pathogen responses.
Keywords: DNA nanofiber networks; antibiotic resistance; bacteria E. coli; biomimetic materials; neutrophil extracellular traps.
© 2019 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Conflict of Interest
The authors declare no competing interest.
Figures

References
-
- a) Kumar DKV, Choi SH, Washicosky KJ, Eimer WA, Tucker S, Ghofrani J, Lefkowitz A, McColl G, Goldstein LE, Tanzi RE, Moir RD, Sci. Transl. Med 2016, 8, 340ra72; - PMC - PubMed
- b) Shimanovich U, Efimov I, Mason TO, Flagmeier P, Buell AK, Gedanken A, Linse S, Åkerfeldt KS, Dobson CM, Weitz DA, Knowles TP, ACS nano 2015, 9, 43. - PubMed
-
- Brinkmann V, Zychlinsky A, Nat. Rev. Microbiol 2007, 5, 577–582. - PubMed
MeSH terms
Substances
Grants and funding
- R01 GM123517/GM/NIGMS NIH HHS/United States
- K08 AI128006/AI/NIAID NIH HHS/United States
- I01 BX002788/BX/BLRD VA/United States
- U01 CA210152/NH/NIH HHS/United States
- R01 GM123517/NH/NIH HHS/United States
- R01 AI141883/NH/NIH HHS/United States
- U01 CA210152/CA/NCI NIH HHS/United States
- R01 HL134846/NH/NIH HHS/United States
- K08 AI128006/NH/NIH HHS/United States
- U19 AI116482/AI/NIAID NIH HHS/United States
- T32 GM145304/GM/NIGMS NIH HHS/United States
- R01 AI141883/AI/NIAID NIH HHS/United States
- K08 AR066569/AR/NIAMS NIH HHS/United States
- R01 HL134846/HL/NHLBI NIH HHS/United States
- U19 AI116482/NH/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

