Network hubs affect evolvability
- PMID: 30699103
- PMCID: PMC6370235
- DOI: 10.1371/journal.pbio.3000111
Network hubs affect evolvability
Abstract
The regulatory processes in cells are typically organized into complex genetic networks. However, it is still unclear how this network structure modulates the evolution of cellular regulation. One would expect that mutations in central and highly connected modules of a network (so-called hubs) would often result in a breakdown and therefore be an evolutionary dead end. However, a new study by Koubkova-Yu and colleagues finds that in some circumstances, altering a hub can offer a quick evolutionary advantage. Specifically, changes in a hub can induce significant phenotypic changes that allow organisms to move away from a local fitness peak, whereas the fitness defects caused by the perturbed hub can be mitigated by mutations in its interaction partners. Together, the results demonstrate how network architecture shapes and facilitates evolutionary adaptation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
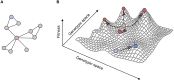
Comment on
-
Heterologous Hsp90 promotes phenotypic diversity through network evolution.PLoS Biol. 2018 Nov 15;16(11):e2006450. doi: 10.1371/journal.pbio.2006450. eCollection 2018 Nov. PLoS Biol. 2018. PMID: 30439936 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

