Oncogenic KSHV-encoded interferon regulatory factor upregulates HMGB2 and CMPK1 expression to promote cell invasion by disrupting a complex lncRNA-OIP5-AS1/miR-218-5p network
- PMID: 30699189
- PMCID: PMC6370251
- DOI: 10.1371/journal.ppat.1007578
Oncogenic KSHV-encoded interferon regulatory factor upregulates HMGB2 and CMPK1 expression to promote cell invasion by disrupting a complex lncRNA-OIP5-AS1/miR-218-5p network
Erratum in
-
Correction: Oncogenic KSHV-encoded interferon regulatory factor upregulates HMGB2 and CMPK1 expression to promote cell invasion by disrupting a complex lncRNA-OIP5-AS1/miR-218-5p network.PLoS Pathog. 2020 Oct 21;16(10):e1009039. doi: 10.1371/journal.ppat.1009039. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33085723 Free PMC article.
Abstract
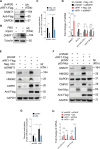
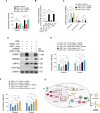
Kaposi's sarcoma (KS), a highly disseminated tumor of hyperproliferative spindle endothelial cells, is the most common AIDS-associated malignancy caused by infection of Kaposi's sarcoma-associated herpesvirus (KSHV). KSHV-encoded viral interferon regulatory factor 1 (vIRF1) is a viral oncogene but its role in KSHV-induced tumor invasiveness and motility remains unknown. Here, we report that vIRF1 promotes endothelial cell migration, invasion and proliferation by down-regulating miR-218-5p to relieve its suppression of downstream targets high mobility group box 2 (HMGB2) and cytidine/uridine monophosphate kinase 1 (CMPK1). Mechanistically, vIRF1 inhibits p53 function to increase the expression of DNA methyltransferase 1 (DNMT1) and DNA methylation of the promoter of pre-miR-218-1, a precursor of miR-218-5p, and increases the expression of a long non-coding RNA OIP5 antisense RNA 1 (lnc-OIP5-AS1), which acts as a competing endogenous RNA (ceRNA) of miR-218-5p to inhibit its function and reduce its stability. Moreover, lnc-OIP5-AS1 increases DNA methylation of the pre-miR-218-1 promoter. Finally, deletion of vIRF1 from the KSHV genome reduces the level of lnc-OIP5-AS1, increases the level of miR-218-5p, and inhibits KSHV-induced invasion. Together, these results define a novel complex lnc-OIP5-AS1/miR-218-5p network hijacked by vIRF1 to promote invasiveness and motility of KSHV-induced tumors.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
CircRNA ARFGEF1 functions as a ceRNA to promote oncogenic KSHV-encoded viral interferon regulatory factor induction of cell invasion and angiogenesis by upregulating glutaredoxin 3.PLoS Pathog. 2021 Feb 4;17(2):e1009294. doi: 10.1371/journal.ppat.1009294. eCollection 2021 Feb. PLoS Pathog. 2021. PMID: 33539420 Free PMC article.
-
Sperm associated antigen 9 promotes oncogenic KSHV-encoded interferon regulatory factor-induced cellular transformation and angiogenesis by activating the JNK/VEGFA pathway.PLoS Pathog. 2020 Aug 10;16(8):e1008730. doi: 10.1371/journal.ppat.1008730. eCollection 2020 Aug. PLoS Pathog. 2020. PMID: 32776977 Free PMC article.
-
An oncogenic viral interferon regulatory factor upregulates CUB domain-containing protein 1 to promote angiogenesis by hijacking transcription factor lymphoid enhancer-binding factor 1 and metastasis suppressor CD82.Cell Death Differ. 2020 Dec;27(12):3289-3306. doi: 10.1038/s41418-020-0578-0. Epub 2020 Jun 17. Cell Death Differ. 2020. PMID: 32555380 Free PMC article.
-
A review on the role of oncogenic lncRNA OIP5-AS1 in human malignancies.Biomed Pharmacother. 2021 May;137:111366. doi: 10.1016/j.biopha.2021.111366. Epub 2021 Feb 15. Biomed Pharmacother. 2021. PMID: 33601149 Review.
-
Distinct roles of Kaposi's sarcoma-associated herpesvirus-encoded viral interferon regulatory factors in inflammatory response and cancer.J Virol. 2013 Sep;87(17):9398-410. doi: 10.1128/JVI.03315-12. Epub 2013 Jun 19. J Virol. 2013. PMID: 23785197 Free PMC article. Review.
Cited by
-
A viral interferon regulatory factor degrades RNA-binding protein hnRNP Q1 to enhance aerobic glycolysis via recruiting E3 ubiquitin ligase KLHL3 and decaying GDPD1 mRNA.Cell Death Differ. 2022 Nov;29(11):2233-2246. doi: 10.1038/s41418-022-01011-1. Epub 2022 May 10. Cell Death Differ. 2022. PMID: 35538151 Free PMC article.
-
Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery.Nat Rev Mol Cell Biol. 2020 Mar;21(3):123-136. doi: 10.1038/s41580-019-0209-0. Epub 2020 Feb 4. Nat Rev Mol Cell Biol. 2020. PMID: 32020081 Free PMC article. Review.
-
miR-142-5p Inhibits Cell Invasion and Migration by Targeting DNMT1 in Breast Cancer.Oncol Res. 2022 Jan 31;28(9):885-897. doi: 10.3727/096504021X16274672547967. Epub 2021 Jul 28. Oncol Res. 2022. PMID: 34321149 Free PMC article.
-
Systematic analysis of expression profiles of HMGB family members for prognostic application in non-small cell lung cancer.Front Mol Biosci. 2022 Jul 18;9:844618. doi: 10.3389/fmolb.2022.844618. eCollection 2022. Front Mol Biosci. 2022. PMID: 35923467 Free PMC article.
-
Long Non-coding RNAs in Gammaherpesvirus Infections: Their Roles in Tumorigenic Mechanisms.Front Microbiol. 2021 Jan 15;11:604536. doi: 10.3389/fmicb.2020.604536. eCollection 2020. Front Microbiol. 2021. PMID: 33519750 Free PMC article. Review.
References
-
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266(5192):1865–9. Epub 1994/12/16. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

