Dynamic expression and functional analysis of circRNA in granulosa cells during follicular development in chicken
- PMID: 30700247
- PMCID: PMC6354403
- DOI: 10.1186/s12864-019-5462-2
Dynamic expression and functional analysis of circRNA in granulosa cells during follicular development in chicken
Abstract
Background: Circular RNA (circRNA) is a type of noncoding RNA involved in a variety of biological processes, especially in post-transcriptional regulation. The granulosa cells of follicles play a determining role in ovarian development. However, the function of circRNA in chicken follicles is unclear. To better understand the molecular mechanism underlying follicular development and granulosa cell function, we performed a strategy of second-generation sequencing and linear RNA depletion for granulosa cells from small yellow follicles (SYF, 5-8 mm), the smallest hierarchal follicles (F6, 9-12 mm), and the largest hierarchal follicles (F1, ~ 40 mm).
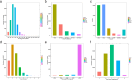
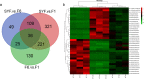
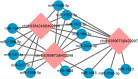
Results: We predicted a total of 11,642 circRNAs that distributed on almost all chromosomes. The majority of the splice lengths of circRNAs were 200-500 nt and mainly produced from intron and CDS regions. During follicle growth, differentially expressed (DE) circRNAs showed dynamic changes which were tissue- and stage-specific. The host genes of DE circRNAs were functionally enriched in GTPase activity and several pathways involved in reproduction. Moreover, bioinformatic prediction analysis for circRalGPS2 demonstrated that circRNAs from the same genes may share common miRNA to act as a sponge. The predicted target genes were enriched in various biological processes including cognition, cell communication, and regulation of signaling, and several pathways related to reproduction such as tight junction, oocyte meiosis, progesterone-mediated oocyte maturation, and GnRH signaling.
Conclusions: This study provides a starting point for further experimental investigations into chicken circRNAs and casts a light on the understanding of follicle development.
Keywords: Chicken; Circular RNA; Follicles; Granulosa cells; Pathways.
Conflict of interest statement
Ethics approval and consent to participate
The experimental chickens were from Jiangsu Jinghai Yellow Chick Co. Ltd., located in Nangtong, Jiangsu Province. All experimental procedures were conducted in compliance with Experimental Animals Regulations and all efforts were made to alleviate the suffering of the birds. Ethics approval for this study was granted by the Animal Care Committee of Yangzhou University (Yangzhou, China) with permit number SYXK (Su) 2012–0029 from the Chinese Ministry of Science and Technology.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures







Similar articles
-
Transcriptome Analysis of circRNA and mRNA in Theca Cells during Follicular Development in Chickens.Genes (Basel). 2020 Apr 29;11(5):489. doi: 10.3390/genes11050489. Genes (Basel). 2020. PMID: 32365656 Free PMC article.
-
The circular RNA aplacirc_13267 upregulates duck granulosa cell apoptosis by the apla-miR-1-13/THBS1 signaling pathway.J Cell Physiol. 2020 Jul;235(7-8):5750-5763. doi: 10.1002/jcp.29509. Epub 2020 Jan 22. J Cell Physiol. 2020. PMID: 31970783
-
CircGRB14 Inhibits Proliferation and Promotes Apoptosis of Granulosa Cells in Chicken Follicle Selection Through Sponging miR-12264-3p and miR-6660-3p.Int J Mol Sci. 2025 Feb 28;26(5):2214. doi: 10.3390/ijms26052214. Int J Mol Sci. 2025. PMID: 40076832 Free PMC article.
-
Chicken ovarian follicles morphology and growth differentiation factor 9 gene expression in chicken ovarian follicles: review.Heliyon. 2022 Jan 11;8(1):e08742. doi: 10.1016/j.heliyon.2022.e08742. eCollection 2022 Jan. Heliyon. 2022. PMID: 35059524 Free PMC article. Review.
-
The roles of cirRNA in the development of germ cells.Acta Histochem. 2020 Apr;122(3):151506. doi: 10.1016/j.acthis.2020.151506. Epub 2020 Jan 31. Acta Histochem. 2020. PMID: 32008790 Review.
Cited by
-
The mediation of pigeon egg production by regulating the steroid hormone biosynthesis of pigeon ovarian granulosa cells.Poult Sci. 2020 Nov;99(11):6075-6083. doi: 10.1016/j.psj.2020.06.048. Epub 2020 Jul 8. Poult Sci. 2020. PMID: 33142527 Free PMC article.
-
Dynamic Expression and Regulatory Network of Circular RNA for Abdominal Preadipocytes Differentiation in Chicken (Gallus gallus).Front Cell Dev Biol. 2021 Nov 12;9:761638. doi: 10.3389/fcell.2021.761638. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34869349 Free PMC article.
-
Circular RNA profiling in the oocyte and cumulus cells reveals that circARMC4 is essential for porcine oocyte maturation.Aging (Albany NY). 2019 Sep 28;11(18):8015-8034. doi: 10.18632/aging.102315. Epub 2019 Sep 28. Aging (Albany NY). 2019. PMID: 31562810 Free PMC article.
-
CircRALGPS2 modulates chicken follicle development through promotion of granulosa cell apoptosis and autophagy via the miR-200a-3p/TGFβ2/SMAD pathway.Poult Sci. 2025 Aug;104(8):105313. doi: 10.1016/j.psj.2025.105313. Epub 2025 May 17. Poult Sci. 2025. PMID: 40446676 Free PMC article.
-
Comprehensive CircRNA Profiling and Selection of Key CircRNAs Reveal the Potential Regulatory Roles of CircRNAs throughout Ovarian Development and Maturation in Cynoglossus semilaevis.Biology (Basel). 2021 Aug 26;10(9):830. doi: 10.3390/biology10090830. Biology (Basel). 2021. PMID: 34571707 Free PMC article.
References
-
- Bahr JM. The chicken ovary as a model of follicular development. Semin Reprod Med. 1991;9(04):352–359. doi: 10.1055/s-2007-1019427. - DOI
-
- Hocking PM. Biology of breeding poultry. 2009.
-
- Palmer SS, Bahr JM. Follicle stimulating hormone increases serum oestradiol-17 beta concentrations, number of growing follicles and yolk deposition in aging hens (Gallus gallus domesticus) with decreased egg production. Br Poult Sci. 1992;33(2):403–414. doi: 10.1080/00071669208417478. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

