Drosophila Subdued is a moonlighting transmembrane protein 16 (TMEM16) that transports ions and phospholipids
- PMID: 30700552
- PMCID: PMC6433057
- DOI: 10.1074/jbc.AC118.006530
Drosophila Subdued is a moonlighting transmembrane protein 16 (TMEM16) that transports ions and phospholipids
Abstract
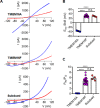
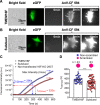
Transmembrane protein 16 (TMEM16) family members play numerous important physiological roles, ranging from controlling membrane excitability and secretion to mediating blood coagulation and viral infection. These diverse functions are largely due to their distinct biophysical properties. Mammalian TMEM16A and TMEM16B are Ca2+-activated Cl- channels (CaCCs), whereas mammalian TMEM16F, fungal afTMEM16, and nhTMEM16 are moonlighting (multifunctional) proteins with both Ca2+-activated phospholipid scramblase (CaPLSase) and Ca2+-activated, nonselective ion channel (CAN) activities. To further understand the biological functions of the enigmatic TMEM16 proteins in different organisms, here, by combining an improved annexin V-based CaPLSase-imaging assay with inside-out patch clamp technique, we thoroughly characterized Subdued, a Drosophila TMEM16 ortholog. We show that Subdued is also a moonlighting transport protein with both CAN and CaPLSase activities. Using a TMEM16F-deficient HEK293T cell line to avoid strong interference from endogenous CaPLSases, our functional characterization and mutagenesis studies revealed that Subdued is a bona fide CaPLSase. Our finding that Subdued is a moonlighting TMEM16 expands our understanding of the molecular mechanisms of TMEM16 proteins and their evolution and physiology in both Drosophila and humans.
Keywords: Anoctamin; CaCC; Drosophila; TMEM16; calcium-binding protein; ion channel; membrane biophysics; membrane transport; phospholipid scramblase; protein moonlighting.
© 2019 Le et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

