Myocardial-specific ablation of Jumonji and AT-rich interaction domain-containing 2 (Jarid2) leads to dilated cardiomyopathy in mice
- PMID: 30700554
- PMCID: PMC6442036
- DOI: 10.1074/jbc.RA118.005634
Myocardial-specific ablation of Jumonji and AT-rich interaction domain-containing 2 (Jarid2) leads to dilated cardiomyopathy in mice
Abstract
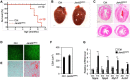
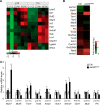
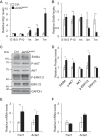
Cardiomyopathy is a common myocardial disease that can lead to sudden death. However, molecular mechanisms underlying cardiomyopathy remain unclear. Jumonji and AT-rich interaction domain-containing 2 (Jarid2) is necessary for embryonic heart development, but functions of Jarid2 after birth remain to be elucidated. Here, we report that myocardial-specific deletion of Jarid2 using αMHC::Cre mice (Jarid2αMHC) causes dilated cardiomyopathy (DCM) and premature death 6-9 months after birth. To determine functions of Jarid2 in the adult heart and DCM, we analyzed gene expression in the heart at postnatal day (p)10 (neonatal) and 7 months (DCM). Pathway analyses revealed that dysregulated genes in Jarid2αMHC hearts at p10, prior to cardiomyopathy, represented heart development and muscle contraction pathways. At 7 months, down-regulated genes in Jarid2αMHC hearts were enriched in metabolic process and ion channel activity pathways and up-regulated genes in extracellular matrix components. In normal hearts, expression levels of contractile genes were increased from p10 to 7 months but were not sufficiently increased in Jarid2αMHC hearts. Moreover, Jarid2 was also necessary to repress fetal contractile genes such as TroponinI1, slow skeletal type (Tnni1) and Actin alpha 2, smooth muscle (Acta2) in neonatal stages through ErbB2-receptor tyrosine kinase 4 (ErbB4) signaling. Interestingly, Ankyrin repeat domain 1 (Ankrd1) and Neuregulin 1 (Nrg1), whose expression levels are known to be increased in the failing heart, were already elevated in Jarid2αMHC hearts within 1 month of birth. Thus, we demonstrate that ablation of Jarid2 in cardiomyocytes results in DCM and suggest that Jarid2 plays important roles in cardiomyocyte maturation during neonatal stages.
Keywords: Cardiomyocyte maturation; Dilated cardiomyopathy; Jarid2; Jumonji family; cardiomyocyte; cardiomyopathy; gene expression; gene regulation; heart failure; muscle contraction.
© 2019 Cho et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures






References
-
- Ujfalusi Z., Vera C. D., Mijailovich S. M., Svicevic M., Yu E. C., Kawana M., Ruppel K. M., Spudich J. A., Geeves M. A., and Leinwand L. A. (2018) Dilated cardiomyopathy myosin mutants have reduced force-generating capacity. J. Biol. Chem. 293, 9017–9029 10.1074/jbc.RA118.001938 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

