The path of pre-ribosomes through the nuclear pore complex revealed by electron tomography
- PMID: 30700705
- PMCID: PMC6353910
- DOI: 10.1038/s41467-019-08342-7
The path of pre-ribosomes through the nuclear pore complex revealed by electron tomography
Abstract
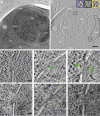
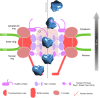
Determining the path of single ribonucleoprotein (RNP) particles through the 100 nm-wide nuclear pore complex (NPC) by fluorescence microscopy remains challenging due to resolution limitation and RNP labeling constraints. By using high-pressure freezing and electron tomography, here we captured snapshots of the translocation of native RNP particles through NPCs in yeast and analyzed their trajectory at nanometer-scale resolution. Morphological and functional analyses indicate that these particles mostly correspond to pre-ribosomes. They are detected in 5-6% of the NPCs, with no apparent bias for NPCs adjacent to the nucleolus. Their path closely follows the central axis of the NPC through the nuclear and inner rings, but diverges at the cytoplasmic ring, suggesting interactions with the cytoplasmic nucleoporins. By applying a probabilistic queueing model to our data, we estimated that the dwell time of pre-ribosomes in the yeast NPC is ~90 ms. These data reveal distinct steps of pre-ribosome translocation through the NPC.
Conflict of interest statement
The authors declare no competing interests.
Figures


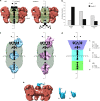

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

