Dual RNA-seq identifies human mucosal immunity protein Mucin-13 as a hallmark of Plasmodium exoerythrocytic infection
- PMID: 30700707
- PMCID: PMC6353872
- DOI: 10.1038/s41467-019-08349-0
Dual RNA-seq identifies human mucosal immunity protein Mucin-13 as a hallmark of Plasmodium exoerythrocytic infection
Abstract
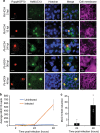
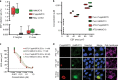
The exoerythrocytic stage of Plasmodium infection is a critical window for prophylactic intervention. Using genome-wide dual RNA sequencing of flow-sorted infected and uninfected hepatoma cells we show that the human mucosal immunity gene, mucin-13 (MUC13), is strongly upregulated during Plasmodium exoerythrocytic hepatic-stage infection. We confirm MUC13 transcript increases in hepatoma cell lines and primary hepatocytes. In immunofluorescence assays, host MUC13 protein expression distinguishes infected cells from adjacent uninfected cells and shows similar colocalization with parasite biomarkers such as UIS4 and HSP70. We further show that localization patterns are species independent, marking both P. berghei and P. vivax infected cells, and that MUC13 can be used to identify compounds that inhibit parasite replication in hepatocytes. This data provides insights into host-parasite interactions in Plasmodium infection, and demonstrates that a component of host mucosal immunity is reprogrammed during the progression of infection.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- W.H.O. World Malaria Report: 2016 (2017).

