Systematic characterization of position one variants within the lantibiotic nisin
- PMID: 30700815
- PMCID: PMC6353901
- DOI: 10.1038/s41598-018-37532-4
Systematic characterization of position one variants within the lantibiotic nisin
Abstract
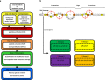
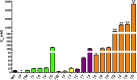
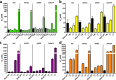
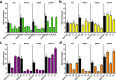
Lantibiotics are a growing class of natural compounds, which possess antimicrobial activity against a broad range of Gram-positive bacteria. Their high potency against human pathogenic strains such as MRSA and VRE makes them excellent candidates as substitutes for classic antibiotics in times of increasing multidrug resistance of bacterial strains. New lantibiotics are detected in genomes and can be heterologously expressed. The functionality of these novel lantibiotics requires a systematic purification and characterization to benchmark them against for example the well-known lantibiotic nisin. Here, we used a standardized workflow to characterize lantibiotics consisting of six individual steps. The expression and secretion of the lantibiotic was performed employing the promiscuous nisin modification machinery. We mutated the first amino acid of nisin into all proteinaceous amino acids and compared their bactericidal potency against sensitive strains as well as strains expressing nisin resistance proteins. Interestingly, we can highlight four distinct groups based on the residual activity of nisin against sensitive as well as resistant L. lactis strains.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Insights in the Antimicrobial Potential of the Natural Nisin Variant Nisin H.Front Microbiol. 2020 Oct 20;11:573614. doi: 10.3389/fmicb.2020.573614. eCollection 2020. Front Microbiol. 2020. PMID: 33193179 Free PMC article.
-
Overexpression, purification and crystallization of the response regulator NsrR involved in nisin resistance.Acta Crystallogr F Struct Biol Commun. 2015 Oct;71(Pt 10):1322-6. doi: 10.1107/S2053230X15016441. Epub 2015 Sep 23. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 26457525 Free PMC article.
-
Genetics of subtilin and nisin biosyntheses: biosynthesis of lantibiotics.Antonie Van Leeuwenhoek. 1996 Feb;69(2):109-17. doi: 10.1007/BF00399416. Antonie Van Leeuwenhoek. 1996. PMID: 8775971 Review.
-
Bypassing lantibiotic resistance by an effective nisin derivative.Bioorg Med Chem. 2019 Aug 1;27(15):3454-3462. doi: 10.1016/j.bmc.2019.06.031. Epub 2019 Jun 20. Bioorg Med Chem. 2019. PMID: 31253534
-
Bioengineering of the model lantibiotic nisin.Bioengineered. 2015;6(4):187-92. doi: 10.1080/21655979.2015.1049781. Epub 2015 May 13. Bioengineered. 2015. PMID: 25970137 Free PMC article. Review.
Cited by
-
Nisin Mutant Prevention Concentration and the Role of Subinhibitory Concentrations on Resistance Development by Diabetic Foot Staphylococci.Antibiotics (Basel). 2022 Jul 19;11(7):972. doi: 10.3390/antibiotics11070972. Antibiotics (Basel). 2022. PMID: 35884226 Free PMC article.
-
Recombinant production of the lantibiotic nisin using Corynebacterium glutamicum in a two-step process.Microb Cell Fact. 2022 Jan 15;21(1):11. doi: 10.1186/s12934-022-01739-y. Microb Cell Fact. 2022. PMID: 35033086 Free PMC article.
-
Therapeutic Application of Lantibiotics and Other Lanthipeptides: Old and New Findings.Appl Environ Microbiol. 2021 Jun 25;87(14):e0018621. doi: 10.1128/AEM.00186-21. Epub 2021 Jun 25. Appl Environ Microbiol. 2021. PMID: 33962984 Free PMC article. Review.
-
Engineered bacterial host for genetic encoding of physiologically stable protein nitration.Front Mol Biosci. 2022 Oct 24;9:992748. doi: 10.3389/fmolb.2022.992748. eCollection 2022. Front Mol Biosci. 2022. PMID: 36353730 Free PMC article.
-
Insights in the Antimicrobial Potential of the Natural Nisin Variant Nisin H.Front Microbiol. 2020 Oct 20;11:573614. doi: 10.3389/fmicb.2020.573614. eCollection 2020. Front Microbiol. 2020. PMID: 33193179 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical

