PU.1 controls fibroblast polarization and tissue fibrosis
- PMID: 30700907
- PMCID: PMC6526281
- DOI: 10.1038/s41586-019-0896-x
PU.1 controls fibroblast polarization and tissue fibrosis
Abstract
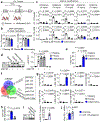
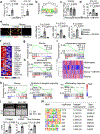
Fibroblasts are polymorphic cells with pleiotropic roles in organ morphogenesis, tissue homeostasis and immune responses. In fibrotic diseases, fibroblasts synthesize abundant amounts of extracellular matrix, which induces scarring and organ failure. By contrast, a hallmark feature of fibroblasts in arthritis is degradation of the extracellular matrix because of the release of metalloproteinases and degrading enzymes, and subsequent tissue destruction. The mechanisms that drive these functionally opposing pro-fibrotic and pro-inflammatory phenotypes of fibroblasts remain unknown. Here we identify the transcription factor PU.1 as an essential regulator of the pro-fibrotic gene expression program. The interplay between transcriptional and post-transcriptional mechanisms that normally control the expression of PU.1 expression is perturbed in various fibrotic diseases, resulting in the upregulation of PU.1, induction of fibrosis-associated gene sets and a phenotypic switch in extracellular matrix-producing pro-fibrotic fibroblasts. By contrast, pharmacological and genetic inactivation of PU.1 disrupts the fibrotic network and enables reprogramming of fibrotic fibroblasts into resting fibroblasts, leading to regression of fibrosis in several organs.
Conflict of interest statement
Conflict of Interest
None
Figures








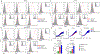



Comment in
-
PU.1 pulls the strings in fibrotic disease.Nat Rev Rheumatol. 2019 Apr;15(4):187. doi: 10.1038/s41584-019-0193-y. Nat Rev Rheumatol. 2019. PMID: 30814690 No abstract available.
-
Recent Insights into the Involvement of Novel Transcription Factors, The Microbiome, and Dysregulated Cellular Metabolism in Pulmonary Fibrosis Pathogenesis.Am J Respir Cell Mol Biol. 2019 Nov;61(5):653-655. doi: 10.1165/rcmb.2019-0196RO. Am J Respir Cell Mol Biol. 2019. PMID: 31265328 Free PMC article. No abstract available.
References
-
- Kalluri R & Zeisberg M Fibroblasts in cancer. Nat Rev Cancer 6, 392–401 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

