CD8+XCR1neg Dendritic Cells Express High Levels of Toll-Like Receptor 5 and a Unique Complement of Endocytic Receptors
- PMID: 30700986
- PMCID: PMC6343586
- DOI: 10.3389/fimmu.2018.02990
CD8+XCR1neg Dendritic Cells Express High Levels of Toll-Like Receptor 5 and a Unique Complement of Endocytic Receptors
Abstract
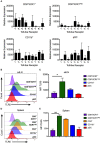
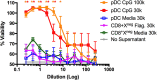
Conventional dendritic cells (cDC) resident in the lymphoid organs of mice have been classically divided into CD8+ and CD8neg subsets. It is well-established that CD8+ dendritic cells (DCs) and their migratory counterparts in the periphery comprise the cross-presenting cDC1 subset. In contrast, CD8neg DCs are grouped together in the heterogeneous cDC2 subset. CD8neg DCs are relatively poor cross-presenters and drive more prominent CD4+ T cell responses against exogenous antigens. The discovery of the X-C motif chemokine receptor 1 (XCR1) as a specific marker of cross-presenting DCs, has led to the identification of a divergent subset of CD8+ DCs that lacks the ability to cross-present. Here, we report that these poorly characterized CD8+XCR1neg DCs have a gene expression profile that is consistent with both plasmacytoid DCs (pDCs) and cDC2. Our data demonstrate that CD8+XCR1neg DCs possess a unique pattern of endocytic receptors and a restricted toll-like receptor (TLR) profile that is particularly enriched for TLR5, giving them a unique position within the DC immunosurveillance network.
Keywords: TLR5; XCR1; dendritic cells; immunosurveillance; microarray; transcriptomics.
Figures





Similar articles
-
Critical roles of a dendritic cell subset expressing a chemokine receptor, XCR1.J Immunol. 2013 Jun 15;190(12):6071-82. doi: 10.4049/jimmunol.1202798. Epub 2013 May 13. J Immunol. 2013. PMID: 23670193
-
Imaging of the cross-presenting dendritic cell subsets in the skin-draining lymph node.Proc Natl Acad Sci U S A. 2016 Jan 26;113(4):1044-9. doi: 10.1073/pnas.1513607113. Epub 2016 Jan 11. Proc Natl Acad Sci U S A. 2016. PMID: 26755602 Free PMC article.
-
A Highly Active Form of XCL1/Lymphotactin Functions as an Effective Adjuvant to Recruit Cross-Presenting Dendritic Cells for Induction of Effector and Memory CD8+ T Cells.Front Immunol. 2018 Nov 27;9:2775. doi: 10.3389/fimmu.2018.02775. eCollection 2018. Front Immunol. 2018. PMID: 30542351 Free PMC article.
-
Conventional Type 1 Dendritic Cells in Intestinal Immune Homeostasis.Front Immunol. 2022 May 27;13:857954. doi: 10.3389/fimmu.2022.857954. eCollection 2022. Front Immunol. 2022. PMID: 35693801 Free PMC article. Review.
-
The CD8+ dendritic cell subset.Immunol Rev. 2010 Mar;234(1):18-31. doi: 10.1111/j.0105-2896.2009.00870.x. Immunol Rev. 2010. PMID: 20193009 Review.
Cited by
-
IFNβ Is a Potent Adjuvant for Cancer Vaccination Strategies.Front Immunol. 2021 Sep 6;12:735133. doi: 10.3389/fimmu.2021.735133. eCollection 2021. Front Immunol. 2021. PMID: 34552594 Free PMC article.
-
Absence of Batf3 reveals a new dimension of cell state heterogeneity within conventional dendritic cells.iScience. 2021 Apr 15;24(5):102402. doi: 10.1016/j.isci.2021.102402. eCollection 2021 May 21. iScience. 2021. PMID: 33997687 Free PMC article.
-
Enhanced Vaccine Immunogenicity Enabled by Targeted Cytosolic Delivery of Tumor Antigens into Dendritic Cells.ACS Cent Sci. 2023 Sep 14;9(9):1835-1845. doi: 10.1021/acscentsci.3c00625. eCollection 2023 Sep 27. ACS Cent Sci. 2023. PMID: 37780364 Free PMC article.
-
The cholera toxin B subunit induces trained immunity in dendritic cells and promotes CD8 T cell antitumor immunity.Front Immunol. 2024 May 15;15:1362289. doi: 10.3389/fimmu.2024.1362289. eCollection 2024. Front Immunol. 2024. PMID: 38812523 Free PMC article.
-
Targeting Xcr1 on Dendritic Cells Rapidly Induce Th1-Associated Immune Responses That Contribute to Protection Against Influenza Infection.Front Immunol. 2022 Feb 28;13:752714. doi: 10.3389/fimmu.2022.752714. eCollection 2022. Front Immunol. 2022. PMID: 35296089 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

