Haplotype analysis of key genes governing grain yield and quality traits across 3K RG panel reveals scope for the development of tailor-made rice with enhanced genetic gains
- PMID: 30701663
- PMCID: PMC6662101
- DOI: 10.1111/pbi.13087
Haplotype analysis of key genes governing grain yield and quality traits across 3K RG panel reveals scope for the development of tailor-made rice with enhanced genetic gains
Abstract
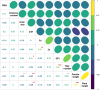
Though several genes governing various major traits have been reported in rice, their superior haplotype combinations for developing ideal variety remains elusive. In this study, haplotype analysis of 120 previously functionally characterized genes, influencing grain yield (87 genes) and grain quality (33 genes) revealed significant variations in the 3K rice genome (RG) panel. For selected genes, meta-expression analysis using already available datasets along with co-expression network provided insights at systems level. Also, we conducted candidate gene based association study for the 120 genes and identified 21 strongly associated genes governing 10-grain yield and quality traits. We report superior haplotypes upon phenotyping the subset of 3K RG panel, SD1-H8 with haplotype frequency (HF) of 30.13% in 3K RG panel, MOC1-H9 (HF: 23.08%), IPA1-H14 (HF: 6.64%), DEP3-H2 (HF: 5.59%), DEP1-H2 (HF: 37.53%), SP1-H3 (HF: 5.05%), LAX1-H5 (HF: 1.56%), LP-H13 (3.64%), OSH1-H4 (5.52%), PHD1-H14 (HF: 15.21%), AGO7-H15 (HF: 3.33%), ROC5-H2 (31.42%), RSR1-H8 (HF: 4.20%) and OsNAS3-H2 (HF: 1.00%). For heading date, Ghd7-H8 (HF: 3.08%), TOB1-H10 (HF: 4.60%) flowered early, Ghd7-H14 (HF: 42.60%), TRX1-H9 (HF: 27.97%), OsVIL3-H14 (HF: 1.72%) for medium duration flowering, while Ghd7-H6 (HF: 1.65%), SNB-H9 (HF: 9.35%) were late flowering. GS5-H4 (HF: 65.84%) attributed slender, GS5-H5 (HF: 29.00%), GW2-H2 (HF: 4.13%) were medium slender and GS5-H9 (HF: 2.15%) for bold grains. Furthermore, haplotype analysis explained possible genetic basis for superiority of selected mega-varieties. Overall, this study suggests the possibility for developing next-generation tailor-made rice with superior haplotype combinations of target genes suiting future food and nutritional demands via haplotype-based breeding.
Keywords: haplotype-based breeding; 3K RG panel; cloned genes; haplotype mining.
© 2019 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The author(s) declare that they have no competing interests.
Figures
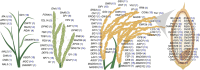


References
-
- Agasimani, S. , Selvakumar, G. , Joel, A.J. and Ganesh Ram, S. (2013) A simple and rapid single kernel screening method to estimate amylose content in rice grains. Phytochem. Anal. 24, 569–573. - PubMed
-
- Ashikari, M. , Sakakibara, H. , Lin, S. , Yamamoto, T. , Takashi, T. , Nishimura, A. , Angeles, E.R. et al. (2005) Cytokinin oxidase regulates rice grain production. Science, 309, 741–745. - PubMed
-
- Bandelt, H.‐J. , Forster, P. and Röhl, A. (1999) Median‐joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. - PubMed
-
- Bevan, M.W. , Uauy, C. , Wulff, B.B. , Zhou, J. , Krasileva, K. and Clark, M.D. (2017) Genomic innovation for crop improvement. Nature, 543, 346. - PubMed
-
- Chandran, A.K.N. , Bhatnagar, N. , Yoo, Y.‐H. , Moon, S. , Park, S.‐A. , Hong, W.‐J. , Kim, B.‐G. et al. (2018) Meta‐expression analysis of unannotated genes in rice and approaches for network construction to suggest the probable roles. Plant Mol. Biol. 96, 17–34. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

