Probable transmission of hepatitis E virus (HEV) via transfusion in the United States
- PMID: 30702157
- PMCID: PMC6501795
- DOI: 10.1111/trf.15140
Probable transmission of hepatitis E virus (HEV) via transfusion in the United States
Abstract
Background: Hepatitis E virus (HEV) can inapparently infect blood donors. To assess transfusion transmission of HEV in the United States, which has not been documented, a donor-recipient repository was evaluated.
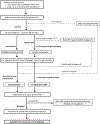
Study design and methods: To identify donations that contained HEV RNA and were linked to patient-recipients with antibody evidence of HEV exposure, we assayed samples from the Retrovirus Epidemiology Donor Study (REDS) Allogeneic Donor and Recipient repository that represents 13,201 linked donations and 3384 transfused patients. Posttransfusion samples, determined to contain IgG anti-HEV by enzyme-linked immunosorbent assay, were reassayed along with corresponding pretransfusion samples for seroconversion (incident exposure) or at least fourfold IgG anti-HEV increase (reexposure). HEV-exposed patients were linked to donations in which HEV RNA was then detected by reverse-transcription quantitative polymerase chain reaction, confirmed by transcription-mediated amplification, and phylogenetically analyzed as subgenomic cDNA sequences.
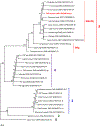
Results: Among all patients, 19 of 1036 (1.8%) who had IgG anti-HEV before transfusion were reexposed; 40 of 2348 (1.7%) without pretransfusion IgG anti-HEV seroconverted. These 59 patients were linked to 257 donations, 1 of which was positive by reverse-transcription quantitative polymerase chain reaction and transcription-mediated amplification. Plasma from this donation contained 5.5 log IU/mL of HEV RNA that grouped with HEV genotype 3, clade 3abchij. The patient-recipient of RBCs from this donation had a greater than eightfold IgG increase; however, clinical data are unavailable.
Conclusions: This is the first report of probable HEV transmission via transfusion in the United States, although it has been frequently observed in Europe and Japan. Additional data on the magnitude of the risk in the United States are needed.
© 2019 AABB.
Conflict of interest statement
Figures
References
-
- Hoofnagle JH, Nelson KE, Purcell RH. Hepatitis E. N.Engl.J.Med. 2012. September 26;367(13):1237–44. - PubMed
-
- Nelson KE, Heaney CD, Kmush BL. The epidemiology and prevention of hepatitis E virus infection. Curr.Epidemiol.Rep. 2017;1–13.
-
- Ankcorn MJ, Tedder RS. Hepatitis E: the current state of play. Transfus.Med. 2017. April 1;27(2):84–95. - PubMed
-
- Boxall E, Herborn A, Kochethu G, Pratt G, Adams D, Ijaz S, Teo CG. Transfusion-transmitted hepatitis E in a ‘nonhyperendemic’ country. Transfus.Med. 2006;16(2):79–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

