Generative modeling of multi-mapping reads with mHi-C advances analysis of Hi-C studies
- PMID: 30702424
- PMCID: PMC6450682
- DOI: 10.7554/eLife.38070
Generative modeling of multi-mapping reads with mHi-C advances analysis of Hi-C studies
Abstract
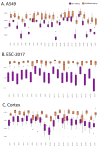
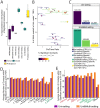
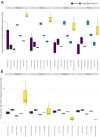
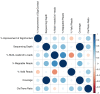
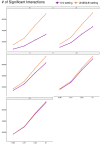
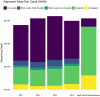
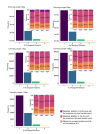
Current Hi-C analysis approaches are unable to account for reads that align to multiple locations, and hence underestimate biological signal from repetitive regions of genomes. We developed and validated mHi-C, a multi-read mapping strategy to probabilistically allocate Hi-C multi-reads. mHi-C exhibited superior performance over utilizing only uni-reads and heuristic approaches aimed at rescuing multi-reads on benchmarks. Specifically, mHi-C increased the sequencing depth by an average of 20% resulting in higher reproducibility of contact matrices and detected interactions across biological replicates. The impact of the multi-reads on the detection of significant interactions is influenced marginally by the relative contribution of multi-reads to the sequencing depth compared to uni-reads, cis-to-trans ratio of contacts, and the broad data quality as reflected by the proportion of mappable reads of datasets. Computational experiments highlighted that in Hi-C studies with short read lengths, mHi-C rescued multi-reads can emulate the effect of longer reads. mHi-C also revealed biologically supported bona fide promoter-enhancer interactions and topologically associating domains involving repetitive genomic regions, thereby unlocking a previously masked portion of the genome for conformation capture studies.
Keywords: Hi-C; chromosome chromatin capture; computational biology; human; mouse; multi-reads; probabilistic modeling; systems biology.
© 2019, Zheng et al.
Conflict of interest statement
YZ, FA, SK No competing interests declared
Figures


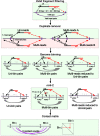




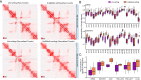





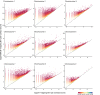
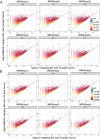







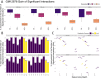


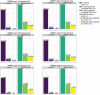
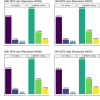
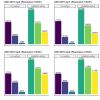


























References
-
- Ay F, Bunnik EM, Varoquaux N, Bol SM, Prudhomme J, Vert JP, Noble WS, Le Roch KG. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Research. 2014b;24:974–988. doi: 10.1101/gr.169417.113. - DOI - PMC - PubMed
-
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Research. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Dryad/10.5061/dryad.v7k3140
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

