Systematic Coarse-Grained Lipid Force Fields with Semiexplicit Solvation via Virtual Sites
- PMID: 30702887
- PMCID: PMC6416712
- DOI: 10.1021/acs.jctc.8b01033
Systematic Coarse-Grained Lipid Force Fields with Semiexplicit Solvation via Virtual Sites
Abstract
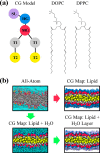
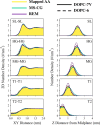
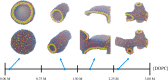
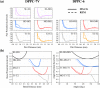
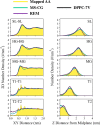
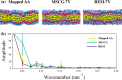
Despite the central role of lipids in many biophysical functions, the molecular mechanisms that dictate macroscopic lipid behavior remain elusive to both experimental and computational approaches. As such, there has been much interest in the development of low-resolution, implicit-solvent coarse-grained (CG) models to dynamically simulate biologically relevant spatiotemporal scales with molecular fidelity. However, in the absence of solvent, a key challenge for CG models is to faithfully emulate solvent-mediated forces, which include both hydrophilic and hydrophobic interactions that drive lipid aggregation and self-assembly. In this work, we provide a new methodological framework to incorporate semiexplicit solvent effects through the use of virtual CG particles, which represent structural features of the solvent-lipid interface. To do so, we leverage two systematic coarse-graining approaches, multiscale coarse-graining (MS-CG) and relative entropy minimization (REM), in a hybrid fashion to construct our virtual-site CG (VCG) models. As a proof-of-concept, we focus our efforts on two lipid species, 1,2-dioleoyl- sn-glycero-3-phosphocholine (DOPC) and 1,2-dipalmitoyl- sn-glycero-3-phosphocholine (DPPC), which adopt a liquid-disordered and gel phase, respectively, at room temperature. Through our analysis, we also present, to our knowledge, the first direct comparison between the MS-CG and REM methods for a complex biomolecule and highlight each of their strengths and weaknesses. We further demonstrate that VCG models recapitulate the rich biophysics of lipids, which enable self-assembly, morphological diversity, and multiple phases. Our findings suggest that the VCG framework is a powerful approach for investigation into macromolecular biophysics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
