EMT network-based feature selection improves prognosis prediction in lung adenocarcinoma
- PMID: 30703089
- PMCID: PMC6354965
- DOI: 10.1371/journal.pone.0204186
EMT network-based feature selection improves prognosis prediction in lung adenocarcinoma
Abstract
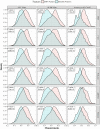
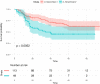
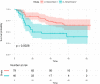
Various feature selection algorithms have been proposed to identify cancer prognostic biomarkers. In recent years, however, their reproducibility is criticized. The performance of feature selection algorithms is shown to be affected by the datasets, underlying networks and evaluation metrics. One of the causes is the curse of dimensionality, which makes it hard to select the features that generalize well on independent data. Even the integration of biological networks does not mitigate this issue because the networks are large and many of their components are not relevant for the phenotype of interest. With the availability of multi-omics data, integrative approaches are being developed to build more robust predictive models. In this scenario, the higher data dimensions create greater challenges. We proposed a phenotype relevant network-based feature selection (PRNFS) framework and demonstrated its advantages in lung cancer prognosis prediction. We constructed cancer prognosis relevant networks based on epithelial mesenchymal transition (EMT) and integrated them with different types of omics data for feature selection. With less than 2.5% of the total dimensionality, we obtained EMT prognostic signatures that achieved remarkable prediction performance (average AUC values >0.8), very significant sample stratifications, and meaningful biological interpretations. In addition to finding EMT signatures from different omics data levels, we combined these single-omics signatures into multi-omics signatures, which improved sample stratifications significantly. Both single- and multi-omics EMT signatures were tested on independent multi-omics lung cancer datasets and significant sample stratifications were obtained.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Lapointe J, Li C, Higgins JP, Van De Rijn M, Bair E, Montgomery K, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(3):811–816. 10.1073/pnas.0304146101 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

