Gene-environment interaction and maternal arsenic methylation efficiency during pregnancy
- PMID: 30703610
- PMCID: PMC7592115
- DOI: 10.1016/j.envint.2019.01.042
Gene-environment interaction and maternal arsenic methylation efficiency during pregnancy
Abstract
Background: Single nucleotide polymorphisms (SNPs) may influence arsenic methylation efficiency, affecting arsenic metabolism. Whether gene-environment interactions affect arsenic metabolism during pregnancy remains unclear, which may have implications for pregnancy outcomes.
Objective: We aimed to investigate main effects as well as potential SNP-arsenic interactions on arsenic methylation efficiency in pregnant women.
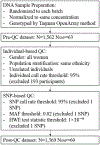
Method: We recruited 1613 pregnant women in Bangladesh, and collected two urine samples from each participant, one at 4-16 weeks, and the second at 21-37 weeks of pregnancy. We determined the proportions of each arsenic metabolite [inorganic As (iAs)%, monomethylarsonic acid (MMA)%, and dimethylarsinic acid (DMA)%] from the total urinary arsenic level of each sample. A panel of 63 candidate SNPs was selected for genotyping based on their reported associations with arsenic metabolism (including in As3MT, N6AMT1, and GSTO2 genes). We used linear regression models to assess the association between each SNP and DMA% with an additive allelic assumption, as well as SNP-arsenic interaction on DMA%. These analyses were performed separately for two urine collection time-points to capture differences in susceptibility to arsenic toxicity.
Result: Intron variants for As3MT were associated with DMA%. rs9527 (β = -2.98%, PFDR = 0.008) and rs1046778 (β = 1.64%, PFDR = 0.008) were associated with this measure in the early gestational period; rs3740393 (β = 2.54%, PFDR = 0.002) and rs1046778 (β = 1.97%, PFDR = 0.003) in the mid-to-late gestational period. Further, As3MT, GSTO2, and N6AMT1 polymorphisms showed different effect sizes on DMA% conditional on arsenic exposure levels. However, SNP-arsenic interactions were not statistically significant after adjusting for false discovery rate (FDR). rs1048546 in N6AMT1 had the highest significance level in the SNP-arsenic interaction test during mid-to-late gestation (β = -1.8% vs. 1.4%, PGxE_FDR = 0.075). Finally, As3MT and As3MT/CNNM2 haplotypes were associated with DMA% at both time points.
Conclusion: We found that not all genetic associations reported in arsenic methylation efficiency replicate in pregnant women. Arsenic exposure level has a limited effect in modifying the association between genetic variation and arsenic methylation efficiency.
Keywords: Arsenic exposure; Arsenic methylation efficiency; Pregnancy; Single nucleotide polymorphism.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Influence of genetic polymorphisms on arsenic methylation efficiency during pregnancy: Evidence from a Spanish birth cohort.Sci Total Environ. 2023 Nov 20;900:165740. doi: 10.1016/j.scitotenv.2023.165740. Epub 2023 Jul 24. Sci Total Environ. 2023. PMID: 37495132
-
Arsenic metabolism, N6AMT1 and AS3MT single nucleotide polymorphisms, and their interaction on gestational diabetes mellitus in Chinese pregnant women.Environ Res. 2023 Mar 15;221:115331. doi: 10.1016/j.envres.2023.115331. Epub 2023 Jan 19. Environ Res. 2023. PMID: 36681142
-
N-6-adenine-specific DNA methyltransferase 1 (N6AMT1) polymorphisms and arsenic methylation in Andean women.Environ Health Perspect. 2013 Jul;121(7):797-803. doi: 10.1289/ehp.1206003. Epub 2013 May 10. Environ Health Perspect. 2013. PMID: 23665909 Free PMC article.
-
AS3MT, GSTO, and PNP polymorphisms: impact on arsenic methylation and implications for disease susceptibility.Environ Res. 2014 Jul;132:156-67. doi: 10.1016/j.envres.2014.03.012. Epub 2014 May 8. Environ Res. 2014. PMID: 24792412 Review.
-
Individual variations in inorganic arsenic metabolism associated with AS3MT genetic polymorphisms.Int J Mol Sci. 2011;12(4):2351-82. doi: 10.3390/ijms12042351. Epub 2011 Apr 4. Int J Mol Sci. 2011. PMID: 21731446 Free PMC article. Review.
Cited by
-
Biomonitoring of inorganic arsenic species in pregnancy.J Expo Sci Environ Epidemiol. 2023 Nov;33(6):921-932. doi: 10.1038/s41370-022-00457-2. Epub 2022 Aug 10. J Expo Sci Environ Epidemiol. 2023. PMID: 35948664 Free PMC article. Review.
-
Association between arsenic (+3 oxidation state) methyltransferase gene polymorphisms and arsenic methylation capacity in rural residents of northern China: a cross-sectional study.Arch Toxicol. 2023 Nov;97(11):2919-2928. doi: 10.1007/s00204-023-03590-5. Epub 2023 Sep 2. Arch Toxicol. 2023. PMID: 37658865
-
Maternal arsenic exposure modifies associations between arsenic, folate and arsenic metabolism gene variants, and spina bifida risk: A case‒control study in Bangladesh.Environ Res. 2024 Nov 15;261:119714. doi: 10.1016/j.envres.2024.119714. Epub 2024 Jul 31. Environ Res. 2024. PMID: 39094898
-
The methyltransferase N6AMT1 participates in the cell cycle by regulating cyclin E levels.PLoS One. 2024 Feb 23;19(2):e0298884. doi: 10.1371/journal.pone.0298884. eCollection 2024. PLoS One. 2024. PMID: 38394175 Free PMC article.
-
[Impact of arsenic exposure on the hepatic metabolic molecular network in obese pregnant mice using metabolomics and proteomics].Se Pu. 2025 Jan;43(1):50-59. doi: 10.3724/SP.J.1123.2024.05028. Se Pu. 2025. PMID: 39722621 Free PMC article. Chinese.
References
-
- Agusa T, Iwata H, Fujihara J, Kunito T, Takeshita H, Minh TB, Trang PT, Viet PH, Tanabe S, 2009. Genetic polymorphisms in AS3MT and arsenic metabolism in residents of the Red River Delta, Vietnam. Toxicol. Appl. Pharmacol 236, 131–141. - PubMed
-
- Ahsan H, Chen Y, Kibriya MG, Slavkovich V, Parvez F, Jasmine F, Gamble MV, Graziano JH, 2007. Arsenic metabolism, genetic susceptibility, and risk of premalignant skin lesions in Bangladesh. Cancer Epidemiol. Biomark. Prev 16, 1270–1278. - PubMed
-
- Antonelli R, Shao K, Thomas DJ, Sams R, Cowden J, 2014. AS3MT, GSTO, and PNP polymorphisms: impact on arsenic methylation and implications for disease susceptibility. Environ. Res 132, 156–167. - PubMed
-
- Argos M, Tong L, Roy S, Sabarinathan M, Ahmed A, Islam MT, Islam T, Rakibuz-Zaman M, Sarwar G, Shahriar H, Rahman M, Yunus M, Graziano JH, Jasmine F, Kibriya MG, Zhou X, Ahsan H, Pierce BL, 2018. Screening for gene-environment (GxE) interaction using omics data from exposed individuals: an application to gene-arsenic interaction. Mamm. Genome 29, 101–111. 10.1007/s00r335-00018-09737-00338. (Epub 02018 Feb 00316). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

