Shared Common Ancestry of Rodent Alphacoronaviruses Sampled Globally
- PMID: 30704076
- PMCID: PMC6409636
- DOI: 10.3390/v11020125
Shared Common Ancestry of Rodent Alphacoronaviruses Sampled Globally
Abstract
The recent discovery of novel alphacoronaviruses (alpha-CoVs) in European and Asian rodents revealed that rodent coronaviruses (CoVs) sampled worldwide formed a discrete phylogenetic group within this genus. To determine the evolutionary history of rodent CoVs in more detail, particularly the relative frequencies of virus-host co-divergence and cross-species transmission, we recovered longer fragments of CoV genomes from previously discovered European rodent alpha-CoVs using a combination of PCR and high-throughput sequencing. Accordingly, the full genome sequence was retrieved from the UK rat coronavirus, along with partial genome sequences from the UK field vole and Poland-resident bank vole CoVs, and a short conserved ORF1b fragment from the French rabbit CoV. Genome and phylogenetic analysis showed that despite their diverse geographic origins, all rodent alpha-CoVs formed a single monophyletic group and shared similar features, such as the same gene constellations, a recombinant beta-CoV spike gene, and similar core transcriptional regulatory sequences (TRS). These data suggest that all rodent alpha CoVs sampled so far originate from a single common ancestor, and that there has likely been a long-term association between alpha CoVs and rodents. Despite this likely antiquity, the phylogenetic pattern of the alpha-CoVs was also suggestive of relatively frequent host-jumping among the different rodent species.
Keywords: alphacoronavirus; ancestry; coronavirus; evolution; recombination; rodents.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

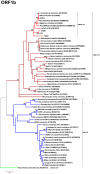
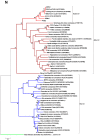


References
-
- Luis A.D., Hayman D.T., O’Shea T.J., Cryan P.M., Gilbert A.T., Pulliam J.R., Mills J.N., Timonin M.E., Willis C.K., Cunningham A.A., et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: Are bats special? Proc. Biol. Sci. 2013;280 doi: 10.1098/rspb.2012.2753. - DOI - PMC - PubMed
-
- Huchon D., Madsen O., Sibbald M.J., Ament K., Stanhope M.J., Catzeflis F., de Jong W.W., Douzery E.J. Rodent phylogeny and a timescale for the evolution of Glires: Evidence from an extensive taxon sampling using three nuclear genes. Mol. Biol. Evol. 2002;19:1053–1065. doi: 10.1093/oxfordjournals.molbev.a004164. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

