Gene editing of the multi-copy H2A.B gene and its importance for fertility
- PMID: 30704500
- PMCID: PMC6357441
- DOI: 10.1186/s13059-019-1633-3
Gene editing of the multi-copy H2A.B gene and its importance for fertility
Abstract
Background: Altering the biochemical makeup of chromatin by the incorporation of histone variants during development represents a key mechanism in regulating gene expression. The histone variant H2A.B, H2A.B.3 in mice, appeared late in evolution and is most highly expressed in the testis. In the mouse, it is encoded by three different genes. H2A.B expression is spatially and temporally regulated during spermatogenesis being most highly expressed in the haploid round spermatid stage. Active genes gain H2A.B where it directly interacts with polymerase II and RNA processing factors within splicing speckles. However, the importance of H2A.B for gene expression and fertility are unknown.
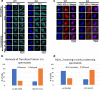
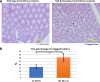
Results: Here, we report the first mouse knockout of this histone variant and its effects on fertility, nuclear organization, and gene expression. In view of the controversy related to the generation of off-target mutations by gene editing approaches, we test the specificity of TALENs by disrupting the H2A.B multi-copy gene family using only one pair of TALENs. We show that TALENs do display a high level of specificity since no off-target mutations are detected by bioinformatics analyses of exome sequences obtained from three consecutive generations of knockout mice and by Sanger DNA sequencing. Male H2A.B.3 knockout mice are subfertile and display an increase in the proportion of abnormal sperm and clogged seminiferous tubules. Significantly, a loss of proper RNA Pol II targeting to distinct transcription-splicing territories and changes to pre-mRNA splicing are observed.
Conclusion: We have produced the first H2A.B knockout mouse using the TALEN approach.
Keywords: Chromatin; Genome editing; H2A.B; Histone variants; Pre-mRNA splicing; RNA polymerase II; Splicing speckles; TALENs.
Conflict of interest statement
Ethics approval and consent to participate
All mouse work was conducted according to protocols approved and authorised by the University of Queensland Animal and the Australian National University Ethics Committee (Protocol A2014/33).
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

