Neuroimaging-Based Classification Algorithm for Predicting 1p/19q-Codeletion Status in IDH-Mutant Lower Grade Gliomas
- PMID: 30705071
- PMCID: PMC7028667
- DOI: 10.3174/ajnr.A5957
Neuroimaging-Based Classification Algorithm for Predicting 1p/19q-Codeletion Status in IDH-Mutant Lower Grade Gliomas
Abstract
Background and purpose: Isocitrate dehydrogenase (IDH)-mutant lower grade gliomas are classified as oligodendrogliomas or diffuse astrocytomas based on 1p/19q-codeletion status. We aimed to test and validate neuroradiologists' performances in predicting the codeletion status of IDH-mutant lower grade gliomas based on simple neuroimaging metrics.
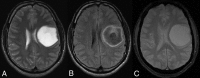
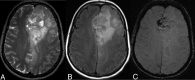
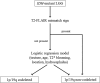
Materials and methods: One hundred two IDH-mutant lower grade gliomas with preoperative MR imaging and known 1p/19q status from The Cancer Genome Atlas composed a training dataset. Two neuroradiologists in consensus analyzed the training dataset for various imaging features: tumor texture, margins, cortical infiltration, T2-FLAIR mismatch, tumor cyst, T2* susceptibility, hydrocephalus, midline shift, maximum dimension, primary lobe, necrosis, enhancement, edema, and gliomatosis. Statistical analysis of the training data produced a multivariate classification model for codeletion prediction based on a subset of MR imaging features and patient age. To validate the classification model, 2 different independent neuroradiologists analyzed a separate cohort of 106 institutional IDH-mutant lower grade gliomas.
Results: Training dataset analysis produced a 2-step classification algorithm with 86.3% codeletion prediction accuracy, based on the following: 1) the presence of the T2-FLAIR mismatch sign, which was 100% predictive of noncodeleted lower grade gliomas, (n = 21); and 2) a logistic regression model based on texture, patient age, T2* susceptibility, primary lobe, and hydrocephalus. Independent validation of the classification algorithm rendered codeletion prediction accuracies of 81.1% and 79.2% in 2 independent readers. The metrics used in the algorithm were associated with moderate-substantial interreader agreement (κ = 0.56-0.79).
Conclusions: We have validated a classification algorithm based on simple, reproducible neuroimaging metrics and patient age that demonstrates a moderate prediction accuracy of 1p/19q-codeletion status among IDH-mutant lower grade gliomas.
© 2019 by American Journal of Neuroradiology.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
