Mutations in the DNMT3A DNA methyltransferase in acute myeloid leukemia patients cause both loss and gain of function and differential regulation by protein partners
- PMID: 30705090
- PMCID: PMC6442042
- DOI: 10.1074/jbc.RA118.006795
Mutations in the DNMT3A DNA methyltransferase in acute myeloid leukemia patients cause both loss and gain of function and differential regulation by protein partners
Abstract
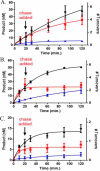
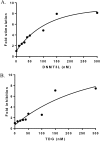
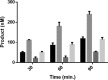
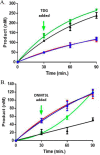
Eukaryotic DNA methylation prevents genomic instability by regulating the expression of oncogenes and tumor-suppressor genes. The negative effects of dysregulated DNA methylation are highlighted by a strong correlation between mutations in the de novo DNA methyltransferase gene DNA methyltransferase 3α (DNMT3A) and poor prognoses among acute myeloid leukemia (AML) patients. We show here that clinically observed DNMT3A mutations dramatically alter enzymatic activity, including mutations that lead to 6-fold hypermethylation and 3-fold hypomethylation of the human cyclin-dependent kinase inhibitor 2B (CDKN2B or p15) gene promoter. Our results provide insights into the clinically observed heterogeneity of p15 methylation in AML. Cytogenetically normal AML (CN-AML) constitutes 40-50% of all AML cases and is the most epigenetically diverse AML subtype with pronounced changes in non-CpG DNA methylation. We identified a subset of DNMT3A mutations that enhance the enzyme's ability to perform non-CpG methylation by 2-8-fold. Many of these mutations mapped to DNMT3A regions known to interact with proteins that themselves contribute to AML, such as thymine DNA glycosylase (TDG). Using functional mapping of TDG-DNMT3A interactions, we provide evidence that TDG and DNMT3-like (DNMT3L) bind distinct regions of DNMT3A. Furthermore, DNMT3A mutations caused diverse changes in the ability of TDG and DNMT3L to affect DNMT3A function. Cell-based studies of one of these DNMT3A mutations (S714C) replicated the enzymatic studies and revealed that it causes dramatic losses of genome-wide methylation. In summary, mutations in DNMT3A lead to diverse levels of activity, interactions with epigenetic machinery components and cellular changes.
Keywords: DNA binding protein; DNA methylation; DNA methyltransferase; enzyme catalysis; enzyme kinetics; enzyme mutation; epigenetics.
© 2019 Sandoval et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


Similar articles
-
p53 and TDG are dominant in regulating the activity of the human de novo DNA methyltransferase DNMT3A on nucleosomes.J Biol Chem. 2021 Jan-Jun;296:100058. doi: 10.1074/jbc.RA120.016125. Epub 2020 Nov 24. J Biol Chem. 2021. PMID: 33172892 Free PMC article.
-
CpG Island Hypermethylation Mediated by DNMT3A Is a Consequence of AML Progression.Cell. 2017 Feb 23;168(5):801-816.e13. doi: 10.1016/j.cell.2017.01.021. Epub 2017 Feb 16. Cell. 2017. PMID: 28215704 Free PMC article.
-
Functional Analysis of DNMT3A DNA Methyltransferase Mutations Reported in Patients with Acute Myeloid Leukemia.Biomolecules. 2019 Dec 18;10(1):8. doi: 10.3390/biom10010008. Biomolecules. 2019. PMID: 31861499 Free PMC article.
-
DNMT3A in Leukemia.Cold Spring Harb Perspect Med. 2017 Feb 1;7(2):a030320. doi: 10.1101/cshperspect.a030320. Cold Spring Harb Perspect Med. 2017. PMID: 28003281 Free PMC article. Review.
-
Significance of targeting DNMT3A mutations in AML.Ann Hematol. 2025 Mar;104(3):1399-1414. doi: 10.1007/s00277-024-05885-8. Epub 2024 Jul 30. Ann Hematol. 2025. PMID: 39078434 Free PMC article. Review.
Cited by
-
Hematopoietic differentiation persists in human iPSCs defective in de novo DNA methylation.BMC Biol. 2022 Jun 15;20(1):141. doi: 10.1186/s12915-022-01343-x. BMC Biol. 2022. PMID: 35705990 Free PMC article.
-
Correlation between the RNA Expression and the DNA Methylation of Estrogen Receptor Genes in Normal and Malignant Human Tissues.Curr Issues Mol Biol. 2024 Apr 19;46(4):3610-3625. doi: 10.3390/cimb46040226. Curr Issues Mol Biol. 2024. PMID: 38666956 Free PMC article.
-
p53 and TDG are dominant in regulating the activity of the human de novo DNA methyltransferase DNMT3A on nucleosomes.J Biol Chem. 2021 Jan-Jun;296:100058. doi: 10.1074/jbc.RA120.016125. Epub 2020 Nov 24. J Biol Chem. 2021. PMID: 33172892 Free PMC article.
-
Active tyrosine phenol-lyase aggregates induced by terminally attached functional peptides in Escherichia coli.J Ind Microbiol Biotechnol. 2020 Aug;47(8):563-571. doi: 10.1007/s10295-020-02294-4. Epub 2020 Jul 31. J Ind Microbiol Biotechnol. 2020. PMID: 32737623 Free PMC article.
-
Probing altered enzyme activity in the biochemical characterization of cancer.Biosci Rep. 2022 Feb 25;42(2):BSR20212002. doi: 10.1042/BSR20212002. Biosci Rep. 2022. PMID: 35048115 Free PMC article. Review.
References
-
- Figueroa M. E., Lugthart S., Li Y., Erpelinck-Verschueren C., Deng X., Christos P. J., Schifano E., Booth J., van Putten W., Skrabanek L., Campagne F., et al. (2010) DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 17, 13–27 10.1016/j.ccr.2009.11.020 - DOI - PMC - PubMed
-
- Melnick A. M. (2010) Epigenetics in AML. Best Pract. Res. Clin. Haematol. 23, 463–468 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

