Plant Extracellular Vesicles Contain Diverse Small RNA Species and Are Enriched in 10- to 17-Nucleotide "Tiny" RNAs
- PMID: 30705133
- PMCID: PMC6447009
- DOI: 10.1105/tpc.18.00872
Plant Extracellular Vesicles Contain Diverse Small RNA Species and Are Enriched in 10- to 17-Nucleotide "Tiny" RNAs
Abstract
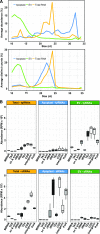
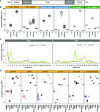
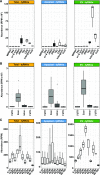
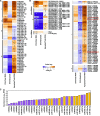
Small RNAs (sRNAs) that are 21 to 24 nucleotides (nt) in length are found in most eukaryotic organisms and regulate numerous biological functions, including transposon silencing, development, reproduction, and stress responses, typically via control of the stability and/or translation of target mRNAs. Major classes of sRNAs in plants include microRNAs (miRNAs) and small interfering RNAs (siRNAs); sRNAs are known to travel as a silencing signal from cell to cell, root to shoot, and even between host and pathogen. In mammals, sRNAs are transported inside extracellular vesicles (EVs), which are mobile membrane-bound compartments that participate in intercellular communication. In addition to sRNAs, EVs carry proteins, lipids, metabolites, and potentially other types of nucleic acids. Here we report that Arabidopsis (Arabidopsis thaliana) EVs also contain diverse species of sRNA. We found that specific miRNAs and siRNAs are preferentially loaded into plant EVs. We also report a previously overlooked class of "tiny RNAs" (10 to 17 nt) that are highly enriched in EVs. This RNA category of unknown function has a broad and very diverse genome origin and might correspond to degradation products.
© 2019 American Society of Plant Biologists. All rights reserved.
Figures

Comment in
-
tyRNA Bubbles: Extracellular Vesicles Carry 10-15-Nucleotide Small RNAs and Specific Groups of MicroRNAs.Plant Cell. 2019 Mar;31(3):558. doi: 10.1105/tpc.19.00130. Epub 2019 Feb 27. Plant Cell. 2019. PMID: 30814256 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

