Force measurements show that uL4 and uL24 mechanically stabilize a fragment of 23S rRNA essential for ribosome assembly
- PMID: 30705137
- PMCID: PMC6426284
- DOI: 10.1261/rna.067504.118
Force measurements show that uL4 and uL24 mechanically stabilize a fragment of 23S rRNA essential for ribosome assembly
Abstract
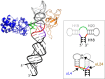
In vitro reconstitution studies have shown that ribosome assembly is highly cooperative and starts with the binding of a few ribosomal (r-) proteins to rRNA. It is unknown how these early binders act. Focusing on the initial stage of the assembly of the large subunit of the Escherichia coli ribosome, we prepared a 79-nucleotide-long region of 23S rRNA encompassing the binding sites of the early binders uL4 and uL24. Force signals were measured in a DNA/RNA dumbbell configuration with a double optical tweezers setup. The rRNA fragment was stretched until unfolded, in the absence or in the presence of the r-proteins (either uL4, uL24, or both). We show that the r-proteins uL4 and uL24 individually stabilize the rRNA fragment, both acting as molecular clamps. Interestingly, this mechanical stabilization is enhanced when both proteins are bound simultaneously. Independently, we observe a cooperative binding of uL4 and uL24 to the rRNA fragment. These two aspects of r-proteins binding both contribute to the efficient stabilization of the 3D structure of the rRNA fragment under investigation. We finally consider implications of our results for large ribosomal subunit assembly.
Keywords: optical trap; ribosome assembly; single molecule.
© 2019 Geffroy et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
