Promoter-proximal pausing mediated by the exon junction complex regulates splicing
- PMID: 30705266
- PMCID: PMC6355915
- DOI: 10.1038/s41467-019-08381-0
Promoter-proximal pausing mediated by the exon junction complex regulates splicing
Abstract
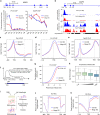
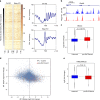
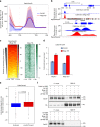
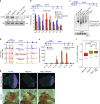
Promoter-proximal pausing of RNA polymerase II (Pol II) is a widespread transcriptional regulatory step across metazoans. Here we find that the nuclear exon junction complex (pre-EJC) is a critical and conserved regulator of this process. Depletion of pre-EJC subunits leads to a global decrease in Pol II pausing and to premature entry into elongation. This effect occurs, at least in part, via non-canonical recruitment of pre-EJC components at promoters. Failure to recruit the pre-EJC at promoters results in increased binding of the positive transcription elongation complex (P-TEFb) and in enhanced Pol II release. Notably, restoring pausing is sufficient to rescue exon skipping and the photoreceptor differentiation defect associated with depletion of pre-EJC components in vivo. We propose that the pre-EJC serves as an early transcriptional checkpoint to prevent premature entry into elongation, ensuring proper recruitment of RNA processing components that are necessary for exon definition.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

