Marine biofilms constitute a bank of hidden microbial diversity and functional potential
- PMID: 30705275
- PMCID: PMC6355793
- DOI: 10.1038/s41467-019-08463-z
Marine biofilms constitute a bank of hidden microbial diversity and functional potential
Abstract
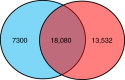
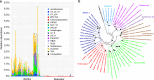
Recent big data analyses have illuminated marine microbial diversity from a global perspective, focusing on planktonic microorganisms. Here, we analyze 2.5 terabases of newly sequenced datasets and the Tara Oceans metagenomes to study the diversity of biofilm-forming marine microorganisms. We identify more than 7,300 biofilm-forming 'species' that are undetected in seawater analyses, increasing the known microbial diversity in the oceans by more than 20%, and provide evidence for differentiation across oceanic niches. Generation of a gene distribution profile reveals a functional core across the biofilms, comprised of genes from a variety of microbial phyla that may play roles in stress responses and microbe-microbe interactions. Analysis of 479 genomes reconstructed from the biofilm metagenomes reveals novel biosynthetic gene clusters and CRISPR-Cas systems. Our data highlight the previously underestimated ocean microbial diversity, and allow mining novel microbial lineages and gene resources.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Marine biofilms: cyanobacteria factories for the global oceans.mSystems. 2024 Nov 19;9(11):e0031724. doi: 10.1128/msystems.00317-24. Epub 2024 Oct 15. mSystems. 2024. PMID: 39404262 Free PMC article.
-
Metagenomic Analysis of Zinc Surface-Associated Marine Biofilms.Microb Ecol. 2019 Feb;77(2):406-416. doi: 10.1007/s00248-018-01313-3. Epub 2019 Jan 5. Microb Ecol. 2019. PMID: 30612183
-
Global marine microbial diversity and its potential in bioprospecting.Nature. 2024 Sep;633(8029):371-379. doi: 10.1038/s41586-024-07891-2. Epub 2024 Sep 4. Nature. 2024. PMID: 39232160 Free PMC article.
-
Unlocking the diversity and biotechnological potential of marine surface associated microbial communities.Curr Opin Microbiol. 2008 Jun;11(3):219-25. doi: 10.1016/j.mib.2008.04.001. Epub 2008 Jun 2. Curr Opin Microbiol. 2008. PMID: 18524668 Review.
-
The Landscape of Global Ocean Microbiome: From Bacterioplankton to Biofilms.Int J Mol Sci. 2023 Mar 30;24(7):6491. doi: 10.3390/ijms24076491. Int J Mol Sci. 2023. PMID: 37047466 Free PMC article. Review.
Cited by
-
Clustered Regularly Interspaced Short Palindromic Repeat/CRISPR-Associated Protein and Its Utility All at Sea: Status, Challenges, and Prospects.Microorganisms. 2024 Jan 6;12(1):118. doi: 10.3390/microorganisms12010118. Microorganisms. 2024. PMID: 38257946 Free PMC article. Review.
-
Mining the Yucatan Coastal Microbiome for the Identification of Non-Ribosomal Peptides Synthetase (NRPS) Genes.Toxins (Basel). 2020 May 26;12(6):349. doi: 10.3390/toxins12060349. Toxins (Basel). 2020. PMID: 32466531 Free PMC article.
-
Genomic and transcriptomic insights into complex virus-prokaryote interactions in marine biofilms.ISME J. 2023 Dec;17(12):2303-2312. doi: 10.1038/s41396-023-01546-2. Epub 2023 Oct 24. ISME J. 2023. PMID: 37875603 Free PMC article.
-
Biotechnological applications of marine bacteria in bioremediation of environments polluted with hydrocarbons and plastics.Appl Microbiol Biotechnol. 2021 Oct;105(19):7171-7185. doi: 10.1007/s00253-021-11569-4. Epub 2021 Sep 13. Appl Microbiol Biotechnol. 2021. PMID: 34515846 Review.
-
Elastic and Self-Healing Copolymer Coatings with Antimicrobial Function.ACS Appl Mater Interfaces. 2024 May 15;16(19):25194-25209. doi: 10.1021/acsami.4c00431. Epub 2024 Apr 29. ACS Appl Mater Interfaces. 2024. PMID: 38684227 Free PMC article.
References
-
- Salta M, Wharton JA, Blache Y, Stokes K, Briand JF. Marine biofilms on artificial surfaces: structure and dynamics. Environ. Microbiol. 2013;15:2879–2893. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

