Concomitant Enhancement of HIV-1 Replication Potential and Neutralization-Resistance in Concert With Three Adaptive Mutations in Env V1/C2/C4 Domains
- PMID: 30705669
- PMCID: PMC6344430
- DOI: 10.3389/fmicb.2019.00002
Concomitant Enhancement of HIV-1 Replication Potential and Neutralization-Resistance in Concert With Three Adaptive Mutations in Env V1/C2/C4 Domains
Abstract
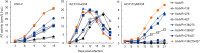
HIV-1 Env protein functions in the entry process and is the target of neutralizing antibodies. Its intrinsically high mutation rate is certainly one of driving forces for persistence/survival in hosts. For optimal replication in various environments, HIV-1 Env must continue to adapt and evolve through balancing sometimes incompatible function, replication fitness, and neutralization sensitivity. We have previously reported that adapted viruses emerge in repeated and prolonged cultures of cells originally infected with a macaque-tropic HIV-1NL4-3 derivative. We have also shown that the adapted viral clones exhibit enhanced growth potentials both in macaque PBMCs and individuals, and that three single-amino acid mutations are present in their Env V1/C2/C4 domains. In this study, we investigated how lab-adapted and highly neutralization-sensitive HIV-1NL4-3 adapts its Env to macaque cells with strongly replication-restrictive nature for HIV-1. While a single and two mutations gave a significantly enhanced replication phenotype in a macaque cell line and also in human cell lines that stably express either human CD4 or macaque CD4, the virus simultaneously carrying the three adaptive mutations always grew best. Entry kinetics of parental and triple mutant viruses were similar, whereas the mutant was significantly more readily inhibited for its infectivity by soluble CD4 than parental virus. Furthermore, molecular dynamics simulations of the Env ectodomain (gp120 and gp41 ectodomain) bound with CD4 suggest that the three mutations increase binding affinity of Env for CD4 in solution. Thus, it is quite likely that the affinity for CD4 of the mutant Env is enhanced relative to the parental Env. Neutralization sensitivity of the triple mutant to CD4 binding site antibodies was not significantly different from that of parental virus, whereas the mutant exhibited a considerably higher resistance against neutralization by a CD4-induced epitope antibody and Env trimer-targeting V1/V2 antibodies. These results suggest that the three adaptive mutations cooperatively promote viral growth via increased CD4 affinity, and also that they enhance viral resistance to several neutralization antibodies by changing the Env-trimer conformation. In total, we have verified here an HIV-1 adaptation pathway in host cells and individuals involving Env derived from a lab-adapted and highly neutralization-sensitive clone.
Keywords: CD4; Env; Env structure; HIV-1; adaptive mutation; neutralization sensitivity; replication potential.
Figures
Similar articles
-
Phenotypic Correlates of HIV-1 Macrophage Tropism.J Virol. 2015 Nov;89(22):11294-311. doi: 10.1128/JVI.00946-15. Epub 2015 Sep 2. J Virol. 2015. PMID: 26339058 Free PMC article.
-
Mutations in HIV-1 envelope that enhance entry with the macaque CD4 receptor alter antibody recognition by disrupting quaternary interactions within the trimer.J Virol. 2015 Jan 15;89(2):894-907. doi: 10.1128/JVI.02680-14. Epub 2014 Nov 5. J Virol. 2015. PMID: 25378497 Free PMC article.
-
Identification of Novel Structural Determinants in MW965 Env That Regulate the Neutralization Phenotype and Conformational Masking Potential of Primary HIV-1 Isolates.J Virol. 2018 Feb 12;92(5):e01779-17. doi: 10.1128/JVI.01779-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237828 Free PMC article.
-
Activation and Inactivation of Primary Human Immunodeficiency Virus Envelope Glycoprotein Trimers by CD4-Mimetic Compounds.J Virol. 2017 Jan 18;91(3):e01880-16. doi: 10.1128/JVI.01880-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881646 Free PMC article.
-
Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41.PLoS Med. 2008 Jan 3;5(1):e9. doi: 10.1371/journal.pmed.0050009. PLoS Med. 2008. PMID: 18177204 Free PMC article.
Cited by
-
Substitution of gp120 C4 region compensates for V3 loss-of-fitness mutations in HIV-1 CRF01_AE co-receptor switching.Emerg Microbes Infect. 2023 Dec;12(1):e2169196. doi: 10.1080/22221751.2023.2169196. Emerg Microbes Infect. 2023. PMID: 36647730 Free PMC article.
-
Species-Specific Valid Ternary Interactions of HIV-1 Env-gp120, CD4, and CCR5 as Revealed by an Adaptive Single-Amino Acid Substitution at the V3 Loop Tip.J Virol. 2021 Jun 10;95(13):e0217720. doi: 10.1128/JVI.02177-20. Epub 2021 Jun 10. J Virol. 2021. PMID: 33883222 Free PMC article.
-
Grand Challenge in Human/Animal Virology: Unseen, Smallest Replicative Entities Shape the Whole Globe.Front Microbiol. 2020 Mar 18;11:431. doi: 10.3389/fmicb.2020.00431. eCollection 2020. Front Microbiol. 2020. PMID: 32256480 Free PMC article. No abstract available.
-
Involvement of a Rarely Used Splicing SD2b Site in the Regulation of HIV-1 vif mRNA Production as Revealed by a Growth-Adaptive Mutation.Viruses. 2023 Dec 14;15(12):2424. doi: 10.3390/v15122424. Viruses. 2023. PMID: 38140666 Free PMC article.
-
Commentary: Derivation of Simian Tropic HIV-1 Infectious Clone Reveals Virus Adaptation to a New Host.Front Cell Infect Microbiol. 2020 May 15;10:235. doi: 10.3389/fcimb.2020.00235. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32500043 Free PMC article. No abstract available.
References
-
- Beaumont T., van Nuenen A., Broersen S., Blattner W. A., Lukashov V. V., Schuitemaker H. (2001). Reversal of human immunodeficiency virus type 1 IIIB to a neutralization-resistant phenotype in an accidentally infected laboratory worker with a progressive clinical course. J. Virol. 75 2246–2252. 10.1128/JVI.75.5.2246-2252.2001 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

