Multi-Method Characterization of the Human Circulating Microbiome
- PMID: 30705670
- PMCID: PMC6345098
- DOI: 10.3389/fmicb.2018.03266
Multi-Method Characterization of the Human Circulating Microbiome
Abstract
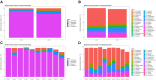
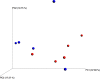
The term microbiome describes the genetic material encoding the various microbial populations that inhabit our body. Whilst colonization of various body niches (e.g., the gut) by dynamic communities of microorganisms is now universally accepted, the existence of microbial populations in other "classically sterile" locations, including the blood, is a relatively new concept. The presence of bacteria-specific DNA in the blood has been reported in the literature for some time, yet the true origin of this is still the subject of much deliberation. The aim of this study was to investigate the phenomenon of a "blood microbiome" by providing a comprehensive description of bacterially derived nucleic acids using a range of complementary molecular and classical microbiological techniques. For this purpose we utilized a set of plasma samples from healthy subjects (n = 5) and asthmatic subjects (n = 5). DNA-level analyses involved the amplification and sequencing of the 16S rRNA gene. RNA-level analyses were based upon the de novo assembly of unmapped mRNA reads and subsequent taxonomic identification. Molecular studies were complemented by viability data from classical aerobic and anaerobic microbial culture experiments. At the phylum level, the blood microbiome was predominated by Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes. The key phyla detected were consistent irrespective of molecular method (DNA vs. RNA), and consistent with the results of other published studies. In silico comparison of our data with that of the Human Microbiome Project revealed that members of the blood microbiome were most likely to have originated from the oral or skin communities. To our surprise, aerobic and anaerobic cultures were positive in eight of out the ten donor samples investigated, and we reflect upon their source. Our data provide further evidence of a core blood microbiome, and provide insight into the potential source of the bacterial DNA/RNA detected in the blood. Further, data reveal the importance of robust experimental procedures, and identify areas for future consideration.
Keywords: biomarker (development); blood microbiome; human; next gen sequencing (NGS); unmapped reads.
Figures




References
LinkOut - more resources
Full Text Sources

