Discovery of a New Natural Product and a Deactivation of a Quorum Sensing System by Culturing a "Producer" Bacterium With a Heat-Killed "Inducer" Culture
- PMID: 30705672
- PMCID: PMC6344404
- DOI: 10.3389/fmicb.2018.03351
Discovery of a New Natural Product and a Deactivation of a Quorum Sensing System by Culturing a "Producer" Bacterium With a Heat-Killed "Inducer" Culture
Abstract
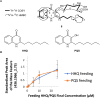
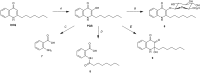
Herein we describe a modified bacterial culture methodology as a tool to discover new natural products via supplementing actinomycete fermentation media with autoclaved cultures of "inducer" microbes. Using seven actinomycetes and four inducer microbes, we detected 28 metabolites that were induced in UHPLC-HRESIMS-based analysis of bacterial fermentations. Metabolomic analysis indicated that each inducer elicited a unique response from the actinomycetes and that some chemical responses were specific to each inducer-producer combination. Among these 28 metabolites, hydrazidomycin D, a new hydrazide-containing natural product was isolated from the pair Streptomyces sp. RKBH-B178 and Mycobacterium smegmatis. This result validated the effectiveness of the strategy in discovering new natural products. From the same set of induced metabolites, an in-depth investigation of a fermentation of Streptomyces sp. RKBH-B178 and autoclaved Pseudomonas aeruginosa led to the discovery of a glucuronidated analog of the pseudomonas quinolone signal (PQS). We demonstrated that RKBH-B178 is able to biotransform the P. aeruginosa quorum sensing molecules, 2-heptyl-4-quinolone (HHQ), and PQS to form PQS-GlcA. Further, PQS-GlcA was shown to have poor binding affinity to PqsR, the innate receptor of HHQ and PQS.
Keywords: biotransformation; induction; metabolomics; natural product discovery; quorum sensing.
Figures



Similar articles
-
A new Pseudomonas quinolone signal (PQS) binding partner: MexG.Chem Sci. 2016 Apr 21;7(4):2553-2562. doi: 10.1039/c5sc04197j. Epub 2016 Jan 20. Chem Sci. 2016. PMID: 28660026 Free PMC article.
-
Interference with Pseudomonas aeruginosa Quorum Sensing and Virulence by the Mycobacterial Pseudomonas Quinolone Signal Dioxygenase AqdC in Combination with the N-Acylhomoserine Lactone Lactonase QsdA.Infect Immun. 2019 Sep 19;87(10):e00278-19. doi: 10.1128/IAI.00278-19. Print 2019 Oct. Infect Immun. 2019. PMID: 31308081 Free PMC article.
-
Development and validation of a UHPLC-MS/MS procedure for quantification of the Pseudomonas Quinolone Signal in bacterial culture after acetylation for characterization of new quorum sensing inhibitors.J Pharm Biomed Anal. 2013 Dec;86:127-34. doi: 10.1016/j.jpba.2013.07.047. Epub 2013 Aug 14. J Pharm Biomed Anal. 2013. PMID: 24001903
-
Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species.Mol Biosyst. 2008 Sep;4(9):882-8. doi: 10.1039/b803796p. Epub 2008 Jun 30. Mol Biosyst. 2008. PMID: 18704225 Review.
-
The third quorum-sensing system of Pseudomonas aeruginosa: Pseudomonas quinolone signal and the enigmatic PqsE protein.J Med Microbiol. 2020 Jan;69(1):25-34. doi: 10.1099/jmm.0.001116. J Med Microbiol. 2020. PMID: 31794380 Review.
Cited by
-
Draft Genome Sequence of Streptomyces sp. Strain RKND-216, an Antibiotic Producer Isolated from Marine Sediment in Prince Edward Island, Canada.Microbiol Resour Announc. 2019 Aug 29;8(35):e00870-19. doi: 10.1128/MRA.00870-19. Microbiol Resour Announc. 2019. PMID: 31467107 Free PMC article.
-
Tryptophan regulates bile and nitrogen metabolism in two pig gut lactobacilli species in vitro based on metabolomics study.Amino Acids. 2022 Oct;54(10):1421-1435. doi: 10.1007/s00726-022-03179-9. Epub 2022 Jul 15. Amino Acids. 2022. PMID: 35838843
-
Antioxidant Activity and Role of Culture Condition in the Optimization of Red Pigment Production by Talaromyces purpureogenus KKP Through Response Surface Methodology.Curr Microbiol. 2020 Aug;77(8):1780-1789. doi: 10.1007/s00284-020-01995-4. Epub 2020 Apr 23. Curr Microbiol. 2020. PMID: 32328751
-
Enhancing chemical and biological diversity by co-cultivation.Front Microbiol. 2023 Feb 1;14:1117559. doi: 10.3389/fmicb.2023.1117559. eCollection 2023. Front Microbiol. 2023. PMID: 36819067 Free PMC article. Review.
-
Insight into the microbial degradation characteristics of polylactic acid by Bacillus sp. JA-4.Arch Microbiol. 2025 Mar 17;207(4):91. doi: 10.1007/s00203-025-04293-4. Arch Microbiol. 2025. PMID: 40097818
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

