Recent Progress on the Molecular Mechanism of Quality Controls Induced by Ribosome Stalling
- PMID: 30705686
- PMCID: PMC6344382
- DOI: 10.3389/fgene.2018.00743
Recent Progress on the Molecular Mechanism of Quality Controls Induced by Ribosome Stalling
Abstract
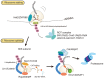
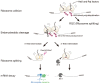
Accurate gene expression is a prerequisite for all cellular processes. Cells actively promote correct protein folding, which prevents the accumulation of abnormal and non-functional proteins. Translation elongation is the fundamental step in gene expression to ensure cellular functions, and abnormal translation arrest is recognized and removed by the quality controls. Recent studies demonstrated that ribosome plays crucial roles as a hub for gene regulation and quality controls. Ribosome-interacting factors are critical for the quality control mechanisms responding to abnormal translation arrest by targeting its products for degradation. Aberrant mRNAs are produced by errors in mRNA maturation steps and cause aberrant translation and are eliminated by the quality control system. In this review, we focus on recent progress on two quality controls, Ribosome-associated Quality Control (RQC) and No-Go Decay (NGD), for abnormal translational elongation. These quality controls recognize aberrant ribosome stalling and induce rapid degradation of aberrant polypeptides and mRNAs thereby maintaining protein homeostasis and preventing the protein aggregation.
Keywords: no-go mRNA decay; ribosome; ribosome stalling; ribosome ubiquitination; ribosome-associated quality control.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

