Emergence of trait variability through the lens of nitrogen assimilation in Prochlorococcus
- PMID: 30706847
- PMCID: PMC6370341
- DOI: 10.7554/eLife.41043
Emergence of trait variability through the lens of nitrogen assimilation in Prochlorococcus
Abstract
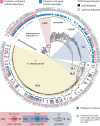
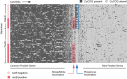
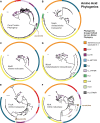
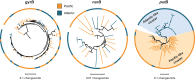
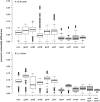
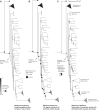
Intraspecific trait variability has important consequences for the function and stability of marine ecosystems. Here we examine variation in the ability to use nitrate across hundreds of Prochlorococcus genomes to better understand the modes of evolution influencing intraspecific allocation of ecologically important functions. Nitrate assimilation genes are absent in basal lineages but occur at an intermediate frequency that is randomly distributed within recently emerged clades. The distribution of nitrate assimilation genes within clades appears largely governed by vertical inheritance, gene loss, and homologous recombination. By mapping this process onto a model of Prochlorococcus' macroevolution, we propose that niche-constructing adaptive radiations and subsequent niche partitioning set the stage for loss of nitrate assimilation genes from basal lineages as they specialized to lower light levels. Retention of these genes in recently emerged lineages has likely been facilitated by selection as they sequentially partitioned into niches where nitrate assimilation conferred a fitness benefit.
Keywords: Cyanobacteria; Prochlorococcus; Synechococcus; ecology; evolutionary biology.
© 2019, Berube et al.
Conflict of interest statement
PB, AR, RB, RS, SC No competing interests declared
Figures







References
-
- Berube PM, Biller SJ, Kent AG, Berta-Thompson JW, Roggensack SE, Roache-Johnson KH, Ackerman M, Moore LR, Meisel JD, Sher D, Thompson LR, Campbell L, Martiny AC, Chisholm SW. Physiology and evolution of nitrate acquisition in Prochlorococcus. The ISME Journal. 2015;9:1195–1207. doi: 10.1038/ismej.2014.211. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

