Activation of MT2 receptor ameliorates dendritic abnormalities in Alzheimer's disease via C/EBPα/miR-125b pathway
- PMID: 30706990
- PMCID: PMC6413662
- DOI: 10.1111/acel.12902
Activation of MT2 receptor ameliorates dendritic abnormalities in Alzheimer's disease via C/EBPα/miR-125b pathway
Abstract
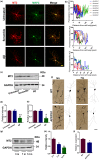
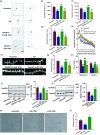
Impairments of dendritic trees and spines have been found in many neurodegenerative diseases, including Alzheimer's disease (AD), in which the deficits of melatonin signal pathway were reported. Melatonin receptor 2 (MT2) is widely expressed in the hippocampus and mediates the biological functions of melatonin. It is known that melatonin application is protective to dendritic abnormalities in AD. However, whether MT2 is involved in the neuroprotection and the underlying mechanisms are not clear. Here, we first found that MT2 is dramatically reduced in the dendritic compartment upon the insult of oligomer Aβ. MT2 activation prevented the Aβ-induced disruption of dendritic complexity and spine. Importantly, activation of MT2 decreased cAMP, which in turn inactivated transcriptional factor CCAAT/enhancer-binding protein α(C/EBPα) to suppress miR-125b expression and elevate the expression of its target, GluN2A. In addition, miR-125b mimics fully blocked the protective effects of MT2 activation on dendritic trees and spines. Finally, injection of a lentivirus containing a miR-125b sponge into the hippocampus of APP/PS1 mice effectively rescued the dendritic abnormalities and learning/memory impairments. Our data demonstrated that the cAMP-C/EBPα/miR-125b/GluN2A signaling pathway is important to the neuroprotective effects of MT2 activation in Aβ-induced dendritic injuries and learning/memory disorders, providing a novel therapeutic target for the treatment of AD synaptopathy.
Keywords: Alzheimer’s disease; MT2; dendritic complexity; dendritic spines; melatonin; miRNA.
© 2019 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Ballard, C. , Gauthier, S. , Corbett, A. , Brayne, C. , Aarsland, D. , & Jones, E. (2011). Alzheimer's disease. Lancet, 377(9770), 1019–1031. - PubMed
-
- Brunner, P. , Sozer‐Topcular, N. , Jockers, R. , Ravid, R. , Angeloni, D. , Fraschini, F. , … Savaskan, E. (2006). Pineal and cortical melatonin receptors MT1 and MT2 are decreased in Alzheimer's disease. European Journal of Histochemistry, 50(4), 311–316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- National Program for Support of Top-Notch Young Professionals/International
- 81829002/National Natural Science Foundation of China/International
- 31721002/National Natural Science Foundation of China/International
- 81761138043/National Natural Science Foundation of China/International
- 91632114/National Natural Science Foundation of China/International
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

