Tandem Affinity Purification and Mass Spectrometry (TAP-MS) for the Analysis of Protein Complexes
- PMID: 30706993
- PMCID: PMC6579647
- DOI: 10.1002/cpps.84
Tandem Affinity Purification and Mass Spectrometry (TAP-MS) for the Analysis of Protein Complexes
Abstract
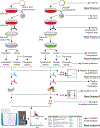
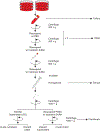
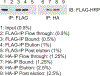
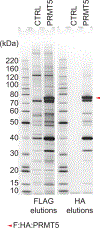
Affinity purification followed by mass spectrometry has become the technique of choice to identify binding partners in biochemical complexes isolated from a physiologic cellular context. In this report we detail our protocol for tandem affinity purification (TAP) primarily based on the use of the FLAG and HA peptide epitopes, with a particular emphasis on factors affecting yield and specificity, as well as steps to implement an automated version of the TAP procedure. © 2019 by John Wiley & Sons, Inc.
Keywords: TAP; affinity purification; epitope tags; mass spectrometry; protein complex; protein-protein interaction.
© 2019 John Wiley & Sons, Inc.
Figures




References
-
- Arsenis C, & McCormick DB (1964). Purification of Liver Flavokinase by Column Chromatography on Flavin-Cellulose Compounds. J Biol Chem, 239, 3093–3097. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

