General functional connectivity: Shared features of resting-state and task fMRI drive reliable and heritable individual differences in functional brain networks
- PMID: 30708106
- PMCID: PMC6462481
- DOI: 10.1016/j.neuroimage.2019.01.068
General functional connectivity: Shared features of resting-state and task fMRI drive reliable and heritable individual differences in functional brain networks
Abstract
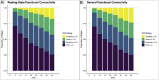
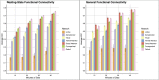
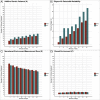
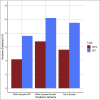
Intrinsic connectivity, measured using resting-state fMRI, has emerged as a fundamental tool in the study of the human brain. However, due to practical limitations, many studies do not collect enough resting-state data to generate reliable measures of intrinsic connectivity necessary for studying individual differences. Here we present general functional connectivity (GFC) as a method for leveraging shared features across resting-state and task fMRI and demonstrate in the Human Connectome Project and the Dunedin Study that GFC offers better test-retest reliability than intrinsic connectivity estimated from the same amount of resting-state data alone. Furthermore, at equivalent scan lengths, GFC displayed higher estimates of heritability than resting-state functional connectivity. We also found that predictions of cognitive ability from GFC generalized across datasets, performing as well or better than resting-state or task data alone. Collectively, our work suggests that GFC can improve the reliability of intrinsic connectivity estimates in existing datasets and, subsequently, the opportunity to identify meaningful correlates of individual differences in behavior. Given that task and resting-state data are often collected together, many researchers can immediately derive more reliable measures of intrinsic connectivity through the adoption of GFC rather than solely using resting-state data. Moreover, by better capturing heritable variation in intrinsic connectivity, GFC represents a novel endophenotype with broad applications in clinical neuroscience and biomarker discovery.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Reliability and reproducibility of individual differences in functional connectivity acquired during task and resting state.Brain Behav. 2016 Mar 30;6(5):e00456. doi: 10.1002/brb3.456. eCollection 2016 May. Brain Behav. 2016. PMID: 27069771 Free PMC article.
-
Boost in Test-Retest Reliability in Resting State fMRI with Predictive Modeling.Cereb Cortex. 2021 May 10;31(6):2822-2833. doi: 10.1093/cercor/bhaa390. Cereb Cortex. 2021. PMID: 33447841 Free PMC article.
-
The Functional Relevance of Task-State Functional Connectivity.J Neurosci. 2021 Mar 24;41(12):2684-2702. doi: 10.1523/JNEUROSCI.1713-20.2021. Epub 2021 Feb 4. J Neurosci. 2021. PMID: 33542083 Free PMC article.
-
What Is the Test-Retest Reliability of Common Task-Functional MRI Measures? New Empirical Evidence and a Meta-Analysis.Psychol Sci. 2020 Jul;31(7):792-806. doi: 10.1177/0956797620916786. Epub 2020 Jun 3. Psychol Sci. 2020. PMID: 32489141 Free PMC article.
-
Can brain state be manipulated to emphasize individual differences in functional connectivity?Neuroimage. 2017 Oct 15;160:140-151. doi: 10.1016/j.neuroimage.2017.03.064. Epub 2017 Mar 31. Neuroimage. 2017. PMID: 28373122 Free PMC article. Review.
Cited by
-
How Tasks Change Whole-Brain Functional Organization to Reveal Brain-Phenotype Relationships.Cell Rep. 2020 Aug 25;32(8):108066. doi: 10.1016/j.celrep.2020.108066. Cell Rep. 2020. PMID: 32846124 Free PMC article.
-
Dopamine-related striatal neurophysiology is associated with specialization of frontostriatal reward circuitry through adolescence.Prog Neurobiol. 2021 Jun;201:101997. doi: 10.1016/j.pneurobio.2021.101997. Epub 2021 Mar 2. Prog Neurobiol. 2021. PMID: 33667595 Free PMC article.
-
The effect of general anesthesia on the test-retest reliability of resting-state fMRI metrics and optimization of scan length.Front Neurosci. 2022 Aug 16;16:937172. doi: 10.3389/fnins.2022.937172. eCollection 2022. Front Neurosci. 2022. PMID: 36051647 Free PMC article.
-
Visual cortical regions show sufficient test-retest reliability while salience regions are unreliable during emotional face processing.Neuroimage. 2020 Oct 15;220:117077. doi: 10.1016/j.neuroimage.2020.117077. Epub 2020 Jun 20. Neuroimage. 2020. PMID: 32574806 Free PMC article.
-
Precision Brain Morphometry Using Cluster Scanning.medRxiv [Preprint]. 2023 Dec 28:2023.12.23.23300492. doi: 10.1101/2023.12.23.23300492. medRxiv. 2023. Update in: Imaging Neurosci (Camb). 2024 May 20;2:imag-2-00175. doi: 10.1162/imag_a_00175. PMID: 38234845 Free PMC article. Updated. Preprint.
References
-
- Aarts A.A., Anderson J.E., Anderson C.J., Attridge P.R., Attwood A., Axt J., Babel M., Bahník Š., Baranski E., Barnett-Cowan M., Bartmess E., Beer J., Bell R., Bentley H., Beyan L., Binion G., Borsboom D., Bosch A., Bosco F.A., Bowman S.D., Brandt M.J., Braswell E., Brohmer H., Brown B.T., Brown K., Brüning J., Calhoun-Sauls A., Callahan S.P., Chagnon E., Chandler J., Chartier C.R., Cheung F., Christopherson C.D., Cillessen L., Clay R., Cleary H., Cloud M.D., Conn M., Cohoon J., Columbus S., Cordes A., Costantini G., Alvarez L.D.C., Cremata E., Crusius J., DeCoster J., DeGaetano M.A., Penna N.D., Den Bezemer B., Deserno M.K., Devitt O., Dewitte L., Dobolyi D.G., Dodson G.T., Donnellan M.B., Donohue R., Dore R.A., Dorrough A., Dreber A., Dugas M., Dunn E.W., Easey K., Eboigbe S., Eggleston C., Embley J., Epskamp S., Errington T.M., Estel V., Farach F.J., Feather J., Fedor A., Fernández-Castilla B., Fiedler S., Field J.G., Fitneva S.A., Flagan T., Forest A.L., Forsell E., Foster J.D., Frank M.C., Frazier R.S., Fuchs H., Gable P., Galak J., Galliani E.M., Gampa A., Garcia S., Gazarian D., Gilbert E., Giner-Sorolla R., Glöckner A., Goellner L., Goh J.X., Goldberg R., Goodbourn P.T., Gordon-McKeon S., Gorges B., Gorges J., Goss J., Graham J., Grange J.A., Gray J., Hartgerink C., Hartshorne J., Hasselman F., Hayes T., Heikensten E., Henninger F., Hodsoll J., Holubar T., Hoogendoorn G., Humphries D.J., Hung C.O.Y., Immelman N., Irsik V.C., Jahn G., Jäkel F., Jekel M., Johannesson M., Johnson L.G., Johnson D.J., Johnson K.M., Johnston W.J., Jonas K., Joy-Gaba J.A., Kappes H.B., Kelso K., Kidwell M.C., Kim S.K., Kirkhart M., Kleinberg B., Knežević G., Kolorz F.M., Kossakowski J.J., Krause R.W., Krijnen J., Kuhlmann T., Kunkels Y.K., Kyc M.M., Lai C.K., Laique A., Lakens D., Lane K.A., Lassetter B., Lazarević L.B., Le Bel E.P., Lee K.J., Lee M., Lemm K., Levitan C.A., Lewis M., Lin L., Lin S., Lippold M., Loureiro D., Luteijn I., MacKinnon S., Mainard H.N., Marigold D.C., Martin D.P., Martinez T., Masicampo E.J., Matacotta J., Mathur M., May M., Mechin N., Mehta P., Meixner J., Melinger A., Miller J.K., Miller M., Moore K., Möschl M., Motyl M., Müller S.M., Munafo M., Neijenhuijs K.I., Nervi T., Nicolas G., Nilsonne G., Nosek B.A., Nuijten M.B., Olsson C., Osborne C., Ostkamp L., Pavel M., Penton-Voak I.S., Perna O., Pernet C., Perugini M., Pipitone R.N., Pitts M., Plessow F., Prenoveau J.M., Rahal R.M., Ratliff K.A., Reinhard D., Renkewitz F., Ricker A.A., Rigney A., Rivers A.M., Roebke M., Rutchick A.M., Ryan R.S., Sahin O., Saide A., Sandstrom G.M., Santos D., Saxe R., Schlegelmilch R., Schmidt K., Scholz S., Seibel L., Selterman D.F., Shaki S., Simpson W.B., Sinclair H.C., Skorinko J.L.M., Slowik A., Snyder J.S., Soderberg C., Sonnleitner C., Spencer N., Spies J.R., Steegen S., Stieger S., Strohminger N., Sullivan G.B., Talhelm T., Tapia M., Te Dorsthorst A., Thomae M., Thomas S.L., Tio P., Traets F., Tsang S., Tuerlinckx F., Turchan P., Valášek M., Van’t Veer A.E., Van Aert R., Van Assen M., Van Bork R., Van De Ven M., Van Den Bergh D., Van Der Hulst M., Van Dooren R., Van Doorn J., Van Renswoude D.R., Van Rijn H., Vanpaemel W., Echeverría A.V., Vazquez M., Velez N., Vermue M., Verschoor M., Vianello M., Voracek M., Vuu G., Wagenmakers E.J., Weerdmeester J., Welsh A., Westgate E.C., Wissink J., Wood M., Woods A., Wright E., Wu S., Zeelenberg M., Zuni K. Estimating the reproducibility of psychological science. Science. 2015;349(80) doi: 10.1126/science.aac4716. - DOI - PubMed
-
- Adams H.H.H., Hibar D.P., Chouraki V., Stein J.L., Nyquist P.A., Rentería M.E., Trompet S., Arias-Vasquez A., Seshadri S., Desrivières S., Beecham A.H., Jahanshad N., Wittfeld K., Van Der Lee S.J., Abramovic L., Alhusaini S., Amin N., Andersson M., Arfanakis K., Aribisala B.S., Armstrong N.J., Athanasiu L., Axelsson T., Beiser A., Bernard M., Bis J.C., Blanken L.M.E., Blanton S.H., Bohlken M.M., Boks M.P., Bralten J., Brickman A.M., Carmichael O., Chakravarty M.M., Chauhan G., Chen Q., Ching C.R.K., Cuellar-Partida G., Braber A. Den, Doan N.T., Ehrlich S., Filippi I., Ge T., Giddaluru S., Goldman A.L., Gottesman R.F., Greven C.U., Grimm O., Griswold M.E., Guadalupe T., Hass J., Haukvik U.K., Hilal S., Hofer E., Hoehn D., Holmes A.J., Hoogman M., Janowitz D., Jia T., Kasperaviciute D., Kim S., Klein M., Kraemer B., Lee P.H., Liao J., Liewald D.C.M., Lopez L.M., Luciano M., Macare C., Marquand A., Matarin M., Mather K.A., Mattheisen M., Mazoyer B., McKay D.R., McWhirter R., Milaneschi Y., Mirza-Schreiber N., Muetzel R.L., Maniega S.M., Nho K., Nugent A.C., Loohuis L.M.O., Oosterlaan J., Papmeyer M., Pappa I., Pirpamer L., Pudas S., Pütz B., Rajan K.B., Ramasamy A., Richards J.S., Risacher S.L., Roiz-Santiañez R., Rommelse N., Rose E.J., Royle N.A., Rundek T., Sämann P.G., Satizabal C.L., Schmaal L., Schork A.J., Shen L., Shin J., Shumskaya E., Smith A.V., Sprooten E., Strike L.T., Teumer A., Thomson R., Tordesillas-Gutierrez D., Toro R., Trabzuni D., Vaidya D., Van Der Grond J., Van Der Meer D., Van Donkelaar M.M.J., Van Eijk K.R., Van Erp T.G.M., Van Rooij D., Walton E., Westlye L.T., Whelan C.D., Windham B.G., Winkler A.M., Woldehawariat G., Wolf C., Wolfers T., Xu B., Yanek L.R., Yang J., Zijdenbos A., Zwiers M.P., Agartz I., Aggarwal N.T., Almasy L., Ames D., Amouyel P., Andreassen O.A., Arepalli S., Assareh A.A., Barral S., Bastin M.E., Becker D.M., Becker J.T., Bennett D.A., Blangero J., Van Bokhoven H., Boomsma D.I., Brodaty H., Brouwer R.M., Brunner H.G., Buckner R.L., Buitelaar J.K., Bulayeva K.B., Cahn W., Calhoun V.D., Cannon D.M., Cavalleri G.L., Chen C., Cheng C.Y., Cichon S., Cookson M.R., Corvin A., Crespo-Facorro B., Curran J.E., Czisch M., Dale A.M., Davies G.E., De Geus E.J.C., De Jager P.L., De Zubicaray G.I., Delanty N., Depondt C., Destefano A.L., Dillman A., Djurovic S., Donohoe G., Drevets W.C., Duggirala R., Dyer T.D., Erk S., Espeseth T., Evans D.A., Fedko I.O., Fernández G., Ferrucci L., Fisher S.E., Fleischman D.A., Ford I., Foroud T.M., Fox P.T., Francks C., Fukunaga M., Gibbs J.R., Glahn D.C., Gollub R.L., Göring H.H.H., Grabe H.J., Green R.C., Gruber O., Gudnason V., Guelfi S., Hansell N.K., Hardy J., Hartman C.A., Hashimoto R., Hegenscheid K., Heinz A., Le Hellard S., Hernandez D.G., Heslenfeld D.J., Ho B.C., Hoekstra P.J., Hoffmann W., Hofman A., Holsboer F., Homuth G., Hosten N., Hottenga J.J., Pol H.E.H., Ikeda M., Ikram M.K., Jack C.R., Jenkinson M., Johnson R., Jönsson E.G., Jukema J.W., Kahn R.S., Kanai R., Kloszewska I., Knopman D.S., Kochunov P., Kwok J.B., Lawrie S.M., Lemaître H., Liu X., Longo D.L., Longstreth W.T., Lopez O.L., Lovestone S., Martinez O., Martinot J.L., Mattay V.S., McDonald C., McIntosh A.M., McMahon K.L., McMahon F.J., Mecocci P., Melle I., Meyer-Lindenberg A., Mohnke S., Montgomery G.W., Morris D.W., Mosley T.H., Mühleisen T.W., Müller-Myhsok B., Nalls M.A., Nauck M., Nichols T.E., Niessen W.J., Nöthen M.M., Nyberg L., Ohi K., Olvera R.L., Ophoff R.A., Pandolfo M., Paus T., Pausova Z., Penninx B.W.J.H., Pike G.B., Potkin S.G., Psaty B.M., Reppermund S., Rietschel M., Roffman J.L., Romanczuk-Seiferth N., Rotter J.I., Ryten M., Sacco R.L., Sachdev P.S., Saykin A.J., Schmidt R., Schofield P.R., Sigurdsson S., Simmons A., Singleton A., Sisodiya S.M., Smith C., Smoller J.W., Soininen H., Srikanth V., Steen V.M., Stott D.J., Sussmann J.E., Thalamuthu A., Tiemeier H., Toga A.W., Traynor B.J., Troncoso J., Turner J.A., Tzourio C., Uitterlinden A.G., Hernández M.C.V., Van Der Brug M., Van Der Lugt A., Van Der Wee N.J.A., Van Duijn C.M., Van Haren N.E.M., Van’t Ent D., Van Tol M.J., Vardarajan B.N., Veltman D.J., Vernooij M.W., Völzke H., Walter H., Wardlaw J.M., Wassink T.H., Weale M.E., Weinberger D.R., Weiner M.W., Wen W., Westman E., White T., Wong T.Y., Wright C.B., Zielke H.R., Zonderman A.B., Deary I.J., Decarli C., Schmidt H., Martin N.G., De Craen A.J.M., Wright M.J., Launer L.J., Schumann G., Fornage M., Franke B., Debette S., Medland S.E., Ikram M.A., Thompson P.M. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat. Neurosci. 2016;19:1569–1582. doi: 10.1038/nn.4398. - DOI - PMC - PubMed
-
- Adhikari B.M., Jahanshad N., Shukla D., Glahn D.C., Blangero J., Fox P.T., Reynolds R.C., Cox R.W., Fieremans E., Veraart J., Novikov D.S., Nichols T.E., Hong L.E., Thompson P.M., Kochunov P. Comparison of heritability estimates on resting state fMRI connectivity phenotypes using the ENIGMA analysis pipeline. Hum. Brain Mapp. 2018 doi: 10.1002/hbm.24331. - DOI - PMC - PubMed
-
- Arfanakis K., Cordes D., Haughton V.M., Moritz C.H., Quigley M.A., Meyerand M.E. Combining independent component analysis and correlation analysis to probe interregional connectivity in fMRI task activation datasets. Magn. Reson. Imaging. 2000;18:921–930. doi: 10.1016/S0730-725X(00)00190-9. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

