An Antigenic Atlas of HIV-1 Escape from Broadly Neutralizing Antibodies Distinguishes Functional and Structural Epitopes
- PMID: 30709739
- PMCID: PMC6435357
- DOI: 10.1016/j.immuni.2018.12.017
An Antigenic Atlas of HIV-1 Escape from Broadly Neutralizing Antibodies Distinguishes Functional and Structural Epitopes
Abstract
Anti-HIV broadly neutralizing antibodies (bnAbs) have revealed vaccine targets on the virus's envelope (Env) protein and are themselves promising immunotherapies. The efficacy of bnAb-based therapies and vaccines depends in part on how readily the virus can escape neutralization. Although structural studies can define contacts between bnAbs and Env, only functional studies can define mutations that confer escape. Here, we mapped how all possible single amino acid mutations in Env affect neutralization of HIV by nine bnAbs targeting five epitopes. For most bnAbs, mutations at only a small fraction of structurally defined contact sites mediated escape, and most escape occurred at sites near, but not in direct contact with, the antibody. The Env mutations selected by two pooled bnAbs were similar to those expected from the combination of the bnAbs's independent action. Overall, our mutation-level antigenic atlas provides a comprehensive dataset for understanding viral immune escape and refining therapies and vaccines.
Keywords: BG505; antibody immunotherapy; deep mutational scanning; immune escape; mutational antigenic profiling; virus evolution.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Competing interests
The authors have no competing interests.
Figures





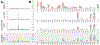
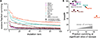
Comment in
-
Playing Chess with HIV.Immunity. 2019 Feb 19;50(2):283-285. doi: 10.1016/j.immuni.2019.01.011. Immunity. 2019. PMID: 30784575
References
-
- Adams RM, Kinney JB, Walczak AM, and Mora T (2017). Physical epistatic landscape of antibody binding affinity 1–15.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

